Endmemo R Tutorial
abs,
acos,
acosh,
addNA,
addTaskCallback,
agrep,
all,
any,
aov,
aperm,
append,
apply,
args,
Array
asinh,
assign,
atan,
atan2,
atanh,
attach,
attachNamespace,
attr,
attributes,
autoload,
backsolve,
Bar Chart Plot
basename,
bessel,
beta,
Binomial Test
body,
Boxplot Example
bquote,
break,
browser,
builtins,
by,
bzfile,
c,
call,
capabilities,
casefold,
cat,
cbind,
ceiling,
char.expand,
character,
charmatch,
charToRaw,
chartr,
Chi Square Test Example
chol,
chol2inv,
choose,
Draw Circle
Object Classes
clipboard,
close,
Clustering Tree Plot
coef,
col,
colMeans,
colnames,
Colors Chart
colSums,
commandArgs,
comment,
complex,
Compress and Decompress
condition handling
conflicts,
Connections
Built-in Constants
contributors,
cos,
cosh,
crossprod,
Cstack_info,
cummax,
cummin,
cumprod,
cumsum,
cut,
data,
Data Frame Data Type
Date and Time, s
debug,
Defunct,
delayedAssign,
density,
deparse,
Deprecated,
det,
dget,
diag,
diff,
difftime,
digamma,
dim,
dimnames,
dir,
dirname,
double,
Quote Text,
drop,
droplevels,
dump,
anyDuplicated,
eapplay,
eigen,
encodeString,
Encoding,
enquote,
environment
Binding and Environment Adjustments
eval,
exists,
exp,
expand.grid,
expm1,
expression,
factor,
factorial,
bzfile,
find.package,
findInterval,
finite,
floor,
flush,
force,
Foreign,
For Loop Example
formals,
format,
forwardsolve,
F-test Example
gamma,
gc,
Garbage Collection
get,
geterrmessage,
getLoadedDLLs,
getNativeSymbolInfo,
getOption,
References to Source Files
gettext,
getwd,
gl,
glm,
gregexpr,
Regular Expression Syntax:
Regular Expression
Function Syntax:
Regular Expression Syntax:
grepl,
Regular Expression Syntax:
gsub,
Regular Expression Syntax:
gzcon,
Heatmap Plot
hexmode,
Histogram Plot Example
I(),
iconv,
icuSetCollate,
identical,
identity,
IF Else Statement
integer,
interaction,
intersect,
intToBits,
intToUtf8,
isSymmetric,
isTRUE,
jitter,
kappa,
kronecker,
l10n_info,
labels,
lapply,
Add Legends to Plot
length,
levels,
library,
Draw Lines
List Data Type
list2env,
load,
log,
log10,
log1p,
log2,
match,
max min,
mean,
Memory
message,
missing,
mode,
name,
names,
nargs,
nchar,
ncol nrow,
noquote,
norm,
Normality Test
normalizePath,
octmode,
open,
Operators
options,
order,
outer,
parse,
Paste
Usage:
Arguments:
Details:
Value:
Plot PCH Symbols Chart
Pie Chart Plot
Plot,
pmatch,
pmax,
pmin,
Draw Points
polyroot,
pretty,
proc.time,
prod,
Colors Chart
Regular Expression
Function Syntax:
Regular Expression Syntax:
Plot PCH Symbols Chart
Plot,
String, s
tapply,
pushBack,
Quantile-Quantile Plot Example
qr,
quit,
Random Number Generation
range,
rank,
raw,
rawConnection,
rbind,
Read.csv Example
Read.delim Example
Read.table Example
regexpr,
Regular Expression Syntax:
remove,
rep,
repeat
replace,
Reserved Words
Trace Copying of Objects
rev,
rle,
row,
sample,
Significance Analysis of Microarrays (samr)
sapply,
save,
saveRDS,
scale,
scan,
Scatter Plots Example
SD SE Calculations
search,
seek,
seq,
sequence,
serialize,
sign,
sink,
solve,
sort,
Matrix
split,
sqrt,
strsplit,
strtoi,
strtrim,
structure,
strwrap,
sub,
Regular Expression Syntax:
subset,
substr,
sum,
summary,
svd,
sweep,
switch,
Sys,
system,
t,
table,
tabulate,
tan,
tanh,
tapply,
tempfile,
textConnection,
tolower,
toString,
toupper,
trace,
transform,
try,
t test
type,
unique,
unlink,
unlist,
unname,
Vector Data Type
version,
warning,
which,
while Loop
wilcoxon rank test
with,
withVisible,
Write Data to File
write.table,
Z-test
abs Function
abs() function computes the absolute value of numeric data.
abs(x)
x: Numeric value, array or vector
> abs(-1)
[1] 1
> abs(20)
[1] 20
> abs(0)
[1] 0
> x <- c(-2,4,0,45,9,-4)
> abs(x)
[1] 2 4 0 45 9 4
> x <- matrix(c(-3,5,-7,1,-9,4),nrow=3,ncol=2,byrow=TRUE)
> abs(x[1,])
[1] 3 5
> abs(x[,1])
[1] 3 7 9
acos Function
acos() function returns the radian arccosine of number data.
acos(x)
x: Numeric value, array or vector
> acos(1)
[1] 0
> acos(0)
[1] 1.570796
> x <- c(-1,-0.866025,-0.707107,-0.5,0,0.5,0.707107,0.866025,1)
> acos(x)
[1] 3.1415926 2.6179931 2.3561948 2.0943951 1.5707963 1.0471976 0.7853979
[8] 0.5235996 0.0000000
acos() function examples list:
acos(x)
(Degrees) |
acos(x)
(Radian) |
x |
| 180 ̊ |
π |
-1 |
| 150 ̊ |
5π/6 |
-0.866025 |
| 135 ̊ |
3π/4 |
-0.707107 |
| 120 ̊ |
2π/3 |
-0.5 |
| 90 ̊ |
π/2 |
0 |
| 60 ̊ |
π/3 |
0.5 |
| 45 ̊ |
π/4 |
0.707107 |
| 30 ̊ |
π/6 |
0.866025 |
| 0 ̊ |
0 |
1 |
acosh Function
acosh() function computes the hyperbolic arccosine of numberic data.
acosh(x)
x: Numeric value, array or vector.
> acosh(1)
[1] 0
> acosh(1.5)
[1] 0.9624237
> x <- c(1,1.5)
> acosh(x)
[1] 0.0000000 0.9624237
addNA Function
addNA() function adds an N/A level of a factor if there are duplications.
addNA(x, ifany=FALSE)
x: vector, factor
> x <- rep(1:10)
> x <- c(x,4,5)
> x
[1] 1 2 3 4 5 6 7 8 9 10 4 5
> factor(x)
[1] 1 2 3 4 5 6 7 8 9 10 4 5
Levels: 1 2 3 4 5 6 7 8 9 10
>addNA(x)
[1] 1 2 3 4 5 6 7 8 9 10 4 5
Levels: 1 2 3 4 5 6 7 8 9 10 <NA>
addTaskCallback Function
addTaskCallback() function registers a R function to be called when a top-level task is completed.
addTaskCallback(f, data=NULL, name=character())
f: the funtion to be added, it has 4 parameters in default
data: the 5th parameter
name: names to be used
times <- function(total = 3, str="Task a") {
ctr <- 0
function(expr, value, ok, visible) {
ctr <<- ctr + 1
cat(str, ctr, "\n")
if(ctr == total) {
cat("handler removing itself\n")
}
return(ctr < total)
}
n <- addTaskCallback(times(4))
removeTaskCallback(n)
agrep Function
agrep() function searches for approximate matches to pattern within each
element of the string.
agrep(pattern, x, ignore.case=FALSE, value=FALSE,
max.distance=0.1, useBytes=FALSE)
pattern: string to be match
x: string vector
ignore.case: if TRUE, ignore case
value: if TRUE, return the matching elements vector, else return the indices vector
...
> x <- c("R language","and","SAND")
> agrep("an",x)
[1] 1 2
> agrep("an",x, ignore.case=TRUE)
[1] 1 2 3
> agrep("uag",x, ignore.case=TRUE)
[1] 1
> agrep("uag",x, ignore.case=TRUE, max=1)
[1] 1
> agrep("uag",x, ignore.case=TRUE, max=2)
[1] 1 2 3
all Function
all() function checks whether all values of a logical vector are true or not.
all(..., na.rm = FALSE)
...: logical vectors
na.rm: if true, NA values are removed
> x <- c(TRUE,TRUE)
> all(x)
[1] TRUE
> x <- c(TRUE,TRUE,FALSE)
> all(x)
[1] FALSE
> x <- c(TRUE,TRUE,NA)
> all(x)
[1] NA
> all(x, na.rm=TRUE)
[1] TRUE
any Function
any() function checks whether there is at one value is true of a logical vector.
any(..., na.rm = FALSE)
...: logical vectors
na.rm: if true, NA values are removed
> x <- c(TRUE,TRUE)
> any(x)
[1] TRUE
> x <- c(TRUE,TRUE,FALSE)
> any(x)
[1] TRUE
> x <- c(TRUE,TRUE,NA)
> any(x)
[1] TRUE
> all(x, na.rm=TRUE)
[1] TRUE
aov Function
aov() function is for analysis of variance (ANOVA).
aov(formula, data=NULL, ...)
formula: a formula specifying the model
data: the data frame containing the variables specified in the formula
Following is a csv file example, we will do ANOVA analysis:
(
Download the data file)
Let first read in the data from the file:
>x <- read.csv("anova.csv",header=T,sep="\t")
One way ANOVA analysis:
> a = aov(Expression~Subtype, data=x)
> summary(a)
Df Sum Sq Mean Sq F value Pr(>F)
Subtype 2 4.75 2.3769 3.991 0.0196 *
Residuals 278 165.59 0.5956
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Please pay attention to the formula format, dependant variance "Expression" is in front of
the independant variance "Subtype".
Report the means and the number of subjects:
>print(model.tables(a,"means"),digits=2)
Tables of means
Grand mean
-0.3053381
Subtype
A B C
-0.18 -0.39 -0.49
rep 143.00 75.00 63.00
Two way ANOVA analysis:
> a = aov(Expression~Subtype*Age, data=x)
> summary(a)
Df Sum Sq Mean Sq F value Pr(>F)
Subtype 2 4.75 2.377 3.975 0.0199 *
Age 1 0.09 0.095 0.159 0.6905
Subtype:Age 2 1.04 0.518 0.866 0.4217
Residuals 275 164.46 0.598
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Here, dependant variance is "Expression", "Subtype" and "Age" are independant variances.
Report the means and the number of subjects:
>print(model.tables(a,"means"),digits=2)
Tables of means
Grand mean
-0.3053381
Gender
f m
-0.39 -0.22
rep 135.00 146.00
Subtype
A B C
-0.22 -0.36 -0.44
rep 143.00 75.00 63.00
Gender:Subtype
Subtype
Gender A B C
f 0 0 -1
rep 40 49 46
m 0 -1 0
rep 103 26 17
aperm Function
aperm() function transposes an array by permuting its dimensions and optionally resizing it.
aperm(x, perm, resize=TRUE, keep.class=TRUE)
x: array
perm: subscript permutation vector
resize: whether array should be resized and elements reordered, default is TRUE
keep.class: whether result should be of the same class of x
...
> x <- array(2:9, c(4,5))
> x
[,1] [,2] [,3] [,4] [,5]
[1,] 2 6 2 6 2
[2,] 3 7 3 7 3
[3,] 4 8 4 8 4
[4,] 5 9 5 9 5
> aperm(x)
[,1] [,2] [,3] [,4]
[1,] 2 3 4 5
[2,] 6 7 8 9
[3,] 2 3 4 5
[4,] 6 7 8 9
[5,] 2 3 4 5
append Function
append() function adds elements to a vector.
append(x, values, after=length(x))
x: vector
values: for appends
after: subscript position which the values are to be appended
...
> x <- rep(1:5)
> x
[1] 1 2 3 4 5
> y <- append(x, 100)
> y
[1] 1 2 3 4 5 100
> y <- append(x, 100, after=2)
> y
[1] 1 2 100 3 4 5
apply Function
apply() function applies a function to margins of an array or matrix.
apply(x,margin,func, ...)
•
x: array
•
margin: subscripts, for matrix, 1 for row, 2 for column
•
func: the function
...
>BOD #R built-in dataset, Biochemical Oxygen Demand
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
Sum up for each row:
> apply(BOD,1,sum)
[1] 9.3 12.3 22.0 20.0 20.6 26.8
Sum up for each column:
> apply(BOD,2,sum)
Time demand
22 89
Multipy all values by 10:
> apply(BOD,1:2,function(x) 10 * x)
Time demand
[1,] 10 83
[2,] 20 103
[3,] 30 190
[4,] 40 160
[5,] 50 156
[6,] 70 198
Used for array, margin set to 1:
> x <- array(1:9)
> apply(x,1,function(x) x * 10)
[1] 10 20 30 40 50 60 70 80 90
Two dimension array, margin can be 1 or 2:
> x <- array(1:9,c(3,3))
> x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> apply(x,1,function(x) x * 10) #or apply(x,2,function(x) x * 10)
[1] 10 20 30 40 50 60 70 80 90
lapply() function can handle data frame with similar results, return is a list:
> lapply(BOD,sum)
$Time
[1] 22
$demand
[1] 89
> lapply(BOD,mean)
$Time
[1] 3.666667
$demand
[1] 14.83333
sapply() has similar function, it defines "simplify=TRUE" by default, thus return a vector:
> sapply(BOD,sum)
Time demand
22 89
> sapply(BOD,sum,simplify=FALSE)
$Time
[1] 22
$demand
[1] 89
args Function
args() function displays the argument names and corresponding default values
of a function or primitive.
args(name)
name: function name
...
> args(append)
function (x, values, after = length(x))
NULL
> args(plot)
function (x, y, ...)
NULL
Array
Array is R data type which has multiple dimensions. array() function creates or tests for arrays. dim() function defines the dimension of an array.
array(data=NA, dim=length(data), dimnames=NULL)
data: vector to fill the array
dim: row and col numbers
:
...
> x <- array(1:9)
> x
[1] 1 2 3 4 5 6 7 8 9
> x <- array(1:9,c(3,3))
> x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> x <- 1:64
> dim(x) <- c(2,4,8) #dim() converts the vector into array
> is.array(x)
[1] TRUE
> x
, , 1
[,1] [,2] [,3] [,4]
[1,] 1 3 5 7
[2,] 2 4 6 8
, , 2
[,1] [,2] [,3] [,4]
[1,] 9 11 13 15
[2,] 10 12 14 16
, , 3
[,1] [,2] [,3] [,4]
[1,] 17 19 21 23
[2,] 18 20 22 24
, , 4
[,1] [,2] [,3] [,4]
[1,] 25 27 29 31
[2,] 26 28 30 32
, , 5
[,1] [,2] [,3] [,4]
[1,] 33 35 37 39
[2,] 34 36 38 40
, , 6
[,1] [,2] [,3] [,4]
[1,] 41 43 45 47
[2,] 42 44 46 48
, , 7
[,1] [,2] [,3] [,4]
[1,] 49 51 53 55
[2,] 50 52 54 56
, , 8
[,1] [,2] [,3] [,4]
[1,] 57 59 61 63
[2,] 58 60 62 64
> x[1,,]
[,1] [,2] [,3] [,4] [,5] [,6] [,7] [,8]
[1,] 1 9 17 25 33 41 49 57
[2,] 3 11 19 27 35 43 51 59
[3,] 5 13 21 29 37 45 53 61
[4,] 7 15 23 31 39 47 55 63
> x[1,2,]
[1] 3 11 19 27 35 43 51 59
> x[1,2,1]
[1] 3
asinh Function
asinh() function computes the hyperbolic arcsine of numberic data.
asinh(x)
x: Numeric value, array or vector.
> asinh(1)
[1] 0.8813736
> asinh(1.5)
[1] 1.194763
> x <- c(1,1.5)
> asinh(x)
[1] 0.8813736 1.1947632
assign Function
assign() function assigns a value to a name in an environment.
assign(x, value, pos = -1, envir = as.environment(pos),
inherits = FALSE, immediate = TRUE)
x: variable name
value: will be assigned to the variable
pos: position to do assignment
envir: the environment to use
...
> assign("z",5)
> z
[1] 5
atan Function
atan() function returns the radian arctangent of number data.
atan(x)
x: Numeric value, array or vector
> atan(1)
[1] 0.7853982
> atan(0)
[1] 0
> atan(0.5)
[1] 0.4636476
> x <- c(1, 0, 0.5)
> atan(x)
[1] 0.7853982 0.0000000 0.4636476
atan2 Function
atan2(y, x) function returns the radian arctangent between the x-axis and the vector from the origin
to (x, y).
atans(y, x)
x, y: Numeric value, array or vector
> atan2(2,1)
[1] 1.107149
> y <- c(2,3)
> x <- c(5,6)
> atan2(y,x)
[1] 0.3805064 0.4636476
atanh Function
atanh() function computes the hyperbolic arctangent of numberic data.
atanh(x)
x: Numeric value, array or vector.
> atanh(0)
[1] 0
> atanh(1)
[1] Inf
> atanh(0.99)
[1] 2.646652
> x <- c(0,1,0.99)
> atanh(x)
[1] 0.000000 Inf 2.646652
attach Function
attach() function makes the data available to the R Search Path.
attach(x)
x: dataframe, matrix, list
Following file has been used for ANOVA analysis:
(
Download the data file)
Let first read in the data from the file:
>x <- read.csv("anova.csv",header=T,sep=",")
There are 3 variables, "Expression", "Gender" and "Subtype". We can display the variables by:
>x$Gender
[1] m m m m m f m m f m m f m m m m f m m m m m m f m m m f m m m m f m m m m
[38] m m m m m m m m m f m f m m m m m f m m f m m f m m m m f m m m m m m m m
[75] m m f m m m m m f m m m m m m m m m f m m f m m f m f m m f m m f m m f m
[112] m f m m f m m m f m m m f m f m f f f f f f m f m f f f m f f f f m f m f
[149] m f f m f f f f f m f m f f m f f m f f m f f f m f f f m f f f m f f m f
[186] f f m f f m f m m f m f m f f m f f f f f m f f m f f f m m m f m m m f f
[223] f f f f f m m m f m f f m f f f m f f f m f f f f m f m f f f f m f f f m
[260] f f m f f f f f f m f f m f f f f f f m f f
Levels: f m
We can't use the variable "Gender" in R Search Path:
>gender
Error: object 'Gender' not found
After attach the object "x", "Gender" can be used globally:
>attach(x)
>Gender
[1] m m m m m f m m f m m f m m m m f m m m m m m f m m m f m m m m f m m m m
[38] m m m m m m m m m f m f m m m m m f m m f m m f m m m m f m m m m m m m m
[75] m m f m m m m m f m m m m m m m m m f m m f m m f m f m m f m m f m m f m
[112] m f m m f m m m f m m m f m f m f f f f f f m f m f f f m f f f f m f m f
[149] m f f m f f f f f m f m f f m f f m f f m f f f m f f f m f f f m f f m f
[186] f f m f f m f m m f m f m f f m f f f f f m f f m f f f m m m f m m m f f
[223] f f f f f m m m f m f f m f f f m f f f m f f f f m f m f f f f m f f f m
[260] f f m f f f f f f m f f m f f f f f f m f f
Levels: f m
detach() function reverses the process:
>detach(x)
>Gender
Error: object 'Gender' not found
attachNamespace Function
attachNamespace() function attaches a namespace to the search path.
attachNamespace(ns, pos=2, dataPath=NULL, depends=NULL)
ns: namespace
pos: position to attach
dataPath: path containing a database of datasets to be lazy-loaded into the attahced
environment
depends: NULL or a character vector of dependencies to be
recorded in object
...
attr Function
attr() function gets or sets specific attributes of an object.
attr(x, which, exact=FALSE)
attr(x, which) <- value
x:
:
:
:
...
attributes Function
attributes() function accesses an object's attributes.
attributes(obj)
attributes(obj) <- value
mostattributes(obj) <- value
obj: object
value: an list of attributes, or NULL
> x <- 3
> attributes(x)
NULL
> x <- matrix(c(3,5,7,1,9,4),nrow=3,ncol=2,byrow=TRUE)
> attributes(x)
$dim
[1] 3 2
> x <- BOD
> x
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
> attributes(x)
$names
[1] "Time" "demand"
$row.names
[1] 1 2 3 4 5 6
$class
[1] "data.frame"
$reference
[1] "A1.4, p. 270"
autoload Function
autoload() function on-demand loads of packages.
autoload(name, package, reset = FALSE, ...)
autoloader(name, package, ...)
name: name of an object
package: name of a package containing the object
...
> require(stats)
> autoload("interpSpline", "splines")
> search()
[1] ".GlobalEnv" "package:stats" "package:graphics"
[4] "package:grDevices" "package:utils" "package:datasets"
[7] "package:methods" "Autoloads" "package:base"
> ls("Autoloads")
[1] "interpSpline"
> .Autoloaded
[1] "splines"
> x <- sort(stats::rnorm(12))
> y <- x^2
> is <- interpSpline(x,y)
> search() #splines loaded
[1] ".GlobalEnv" "package:splines" "package:stats"
[4] "package:graphics" "package:grDevices" "package:utils"
[7] "package:datasets" "package:methods" "Autoloads"
[10] "package:base"
> detach("package:splines")
> search()
[1] ".GlobalEnv" "package:stats" "package:graphics"
[4] "package:grDevices" "package:utils" "package:datasets"
[7] "package:methods" "Autoloads" "package:base"
> is2 <- interpSpline(x,y+x)
> search() #splines loaded
[1] ".GlobalEnv" "package:splines" "package:stats"
[4] "package:graphics" "package:grDevices" "package:utils"
[7] "package:datasets" "package:methods" "Autoloads"
[10] "package:base"
> detach("package:splines")
> search() #splines unloaded
[1] ".GlobalEnv" "package:stats" "package:graphics"
[4] "package:grDevices" "package:utils" "package:datasets"
[7] "package:methods" "Autoloads"
backsolve Function
backsolve() function solves a system of linear
equations where the coefficient matrix is upper triangular.
x <- backsolve (R, b)
backsolve(r, x, k=ncol(r), upper.tri=TRUE, transpose=FALSE)
r: upper triangular matrix
x: a matrix whose columns give the right-hand sides for the equations
k: The number of columns of r and rows of x to use
> r <- rbind(c(1,2,3),c(0,1,1),c(0,0,2))
> y <- backsolve(r, x <- c(8,4,2))
> y
[1] -1 3 1
> r %*% y
[,1]
[1,] 8
[2,] 4
[3,] 2
> backsolve(r, x, transpose = TRUE)
[1] 8 -12 -5
Bar Chart Plot
barplot(...) funtion plot a bar chart. It's usage is:
barplot(height, width = 1, space = NULL,
names.arg = NULL, legend.text = NULL, beside = FALSE,
horiz = FALSE, density = NULL, angle = 45,
col = NULL, border = par("fg"),
main = NULL, sub = NULL, xlab = NULL, ylab = NULL,
xlim = NULL, ylim = NULL, xpd = TRUE, log = "",
axes = TRUE, axisnames = TRUE,
cex.axis = par("cex.axis"), cex.names = par("cex.axis"),
inside = TRUE, plot = TRUE, axis.lty = 0, offset = 0,
add = FALSE, args.legend = NULL, ...)
height: Vector of each bar heights
width: Vector of bar width
space: Space between bars
col: Vector of color for each bar
...
First let's make a simple bar chart:
>x <- c(3,2,6,8,4)
>barplot(x)

Let's add some annotations:
>barplot(x,border="tan2",names.arg=c("Jan","Feb","Mar","Apr","May"),
+ xlab="Month",ylab="Revenue",density=c(0,5,20,50,100))

Suppose the bar chart above is about software department of our company, we are going to
compare other department's revenues including hardware and services:
>A <- matrix(c(3,5,7,1,9,4,6,5,2,12,2,1,7,6,8),nrow=3,ncol=5,byrow=TRUE)
>barplot(A,main="total revenue",names.arg=c("Jan","Feb","Mar","Apr","May"),
+ xlab="month",ylab="revenue",col=c("tan2","blue","darkslategray3"))
>legend(x=0.2,y=24,c("soft","hardware","service"),cex=.8,
+ col=c("tan2","blue","darkslategray3"),pch=c(22,0,0))

Let's compare the data sets horizontally:
>barplot(A,main="total revenue",beside=TRUE,
+ names.arg=c("Jan","Feb","Mar","Apr","May"),
+ xlab="month",ylab="revenue",col=c("tan2","blue","darkslategray3"))
>legend(x=1,y=11,c("soft","hardware","service"),cex=.8,
+ col=c("tan2","blue","darkslategray3"),pch=c(22,0,0))

basename Function
basename() function gets the file name and removes all of the path.
basename(x)
x: path name
> x <- "/usr/local/r/test.R"
> basename(x)
[1] "test.R"
bessel Function
bessel() function computes the bessel function.
besselI(x, nu, expon.scaled = FALSE)
besselK(x, nu, expon.scaled = FALSE)
besselJ(x, nu)
besselY(x, nu)
x: numeric, ≥ 0
nu: numeric; The order (maybe fractional!) of the corresponding Bessel function
expon.scaled: logical; if TRUE, the results are exponentially scaled in order to avoid overflow (I(nu)) or underflow (K(nu)), respectively
beta Function
beta() function return the beta function and the natural logarithm of the beta function.
B(a,b) = Γ(a)Γ(b)/Γ(a+b)
beta(a, b)
lbeta(a, b)
a,b: non-negative numeric vectors
> beta(4,9)
[1] 0.0005050505
> lbeta(4,9)
[1] -7.590852
> x <- c(3,6, 4)
> y <- c(7,4, 12)
> beta(x,y)
[1] 0.0039682540 0.0019841270 0.0001831502
Binomial Test
binom.test() function performs binomial test of null hypothesis about binomial distribution.
binom.test(x,n,p=0.5,alternative=c("two.sided","less","greater"),
conf.level=0.95)
x: number of successes
n: number of trials
p: hypothesized probability of success
alternative: alternative hypothesis, including "two.sided","greater","less"
conf.level: confidence level
Suppose in a coin tossing, the chance to get a head or tail is 50%. In a real case,
we have 100 coin tossings, and get 48 heads, is our original hypothesis true?
> binom.test(48,100)
Exact binomial test
data: 48 and 100
number of successes = 48, number of trials = 100, p-value = 0.7644
alternative hypothesis: true probability of success is not equal to 0.5
95 percent confidence interval:
0.3790055 0.5822102
sample estimates:
probability of success
0.48
Since the p-value is 0.7644, far greater than 0.05, the hypothesis is accepted.
body Function
body() function gets or sets the body of a function.
body(f = sys.function(sys.parent()))
body(f, env = environment(fun)) <- value
f: function object
env: environment of the function
...
> f <- function(x) x^3
> f(3)
[1] 27
> body(f) <- quote(x^2)
> f(3)
[1] 9
Boxplot Example
Boxplot usually refers to box-and-whisker plot, which is a popular method to show data by drawing a box around the 1st and 3rd quartile, and the whiskers for the smallest and largest data values, the median is represented
by a bold line in the box.
Following is a csv file example "boxplot.csv", we will draw a boxplot of "Expression" based on Subtype "A", "B" and "C":
Let first read in the data from the file:
> x <- read.csv("boxplot.csv",header=T,sep="\t")
> x <- t(x)
> a <- as.numeric(x[2,1:143])
> b <- as.numeric(x[2,144:218])
> c <- as.numeric(x[2,219:ncol(x)])
Box plot based on subtype A, B and C:
> boxplot(a,b,c,col=c("red","blue","green"),names=c("A","B","C"),
+ xlab="Subtype",ylab="Expression")

The above plot shows that the Expression values for Subtype A, B and C are similar, however
the two sub-boxes around the median of Subtype C is wider than B and A, the data are
not symmetrically distributed around the median.
if the 'notch' parameter is 'TRUE', a notch is drawn in each side of the boxes. If the notches of two plots do not overlap this is
'strong evidence' that the two medians differ.
> boxplot(a,b,c,col=c("red","blue","green"),names=c("A","B","C"),
+ notch=TRUE, xlab="Subtype",ylab="Expression")

We can write the plot into a file:
> png("boxplot1.png",400,300)
> boxplot(a,b,c,col=c("red","blue","green"),names=c("A","B","C"),
+ xlab="Subtype",ylab="Expression")
> graphics.off()
Boxplot function parameters list:
bquote Function
bquote() function quotes its argument except that terms wrapped in ., and () are evaluated
in the specified environment.
bquote(expr, where = parent.frame())
expr: language object
where: environment
> x <- 5
> bquote(x == x)
x == x
> bquote(x == .(x))
x == 5
> bquote(x == 5)
x == 5
break Function
break() function stops a loop, including for loop, while loop, repeat loop.
> x <- 0
> for (i in 1:10) x <- x + i
> x
[1] 55
> x <- 0
> for (i in 1:10) {if (i == 5) break; x <- x + i}
> x
[1] 10
browser Function
browser() function interrupt the execution of an expression and allow the inspection of the
environment where browser was called from.
browser(text="", condition=NULL, expr=TRUE, skipCalls=0L)
text: a text string that can be retrieved once the browser is invoked
condition: a condition that can be retrieved once the browser is invoked
expr: An expression, which if it evaluates to TRUE the debugger will invoked, otherwise control is returned directly
skipCalls: how many previous calls to skip when reporting the calling context
builtins Function
builtins() function returns the names of all the built-in objects.
builtins(internal = FALSE)
internal: a logical indicating whether only ‘internal’ functions (which can be called via .Internal) should be returned
...
> length(builtins(internal=TRUE))
[1] 492
> length(builtins())
[1] 1269
by Function
by() applies a function to specified subsets of a data frame.
by(data, INDICES, FUN, ..., simplify = TRUE)
•
data: an R object, normally a data frame, possibly a matrix
•
INDICES: a factor or a list of factors, each of length nrow(data)
•
FUN: a function to be applied to data frame subsets of data
...
>Orange #R built-in dataset, Growth of Orange Trees
Tree age circumference
1 1 118 30
2 1 484 58
3 1 664 87
4 1 1004 115
5 1 1231 120
6 1 1372 142
7 1 1582 145
8 2 118 33
9 2 484 69
10 2 664 111
11 2 1004 156
12 2 1231 172
13 2 1372 203
14 2 1582 203
15 3 118 30
16 3 484 51
17 3 664 75
18 3 1004 108
19 3 1231 115
20 3 1372 139
21 3 1582 140
22 4 118 32
23 4 484 62
24 4 664 112
25 4 1004 167
26 4 1231 179
27 4 1372 209
28 4 1582 214
29 5 118 30
30 5 484 49
31 5 664 81
32 5 1004 125
33 5 1231 142
34 5 1372 174
35 5 1582 177
Calculate the mean circumference of different Tree groups:
> x <- by(Orange[,2],Orange[,1],mean)
> x
Orange[, 1]: 3
[1] 922.1429
------------------------------------------------------------
Orange[, 1]: 1
[1] 922.1429
------------------------------------------------------------
Orange[, 1]: 5
[1] 922.1429
------------------------------------------------------------
Orange[, 1]: 2
[1] 922.1429
------------------------------------------------------------
Orange[, 1]: 4
[1] 922.1429
> x[1]
$`3`
[1] 922.1429
> x['3']
$`3`
[1] 922.1429
bzfile Function
bzfile() function open a bzip2-ed file.
bzfile(description, open = "", encoding = getOption("encoding"),
compression = 6)
description: file name or connection.
open: open file mode.
encoding: the name of the encoding to be used.
compression: integer in 0–9. The amount of compression to be applied when writing, from none to maximal available.
...
> writ <- bzfile("tp.bz2", "w") # bzip2-ed file
> cat("writ into bz2 file", "111111111", "", "2222222222",
+ file = writ, sep = "\n")
> close(writ)
> print(readLines(writ <- bzfile("tp.bz2")))
adLines(writ <- bzfile("tp.bz2")))
[1] "writ into bz2 file" "111111111" ""
[4] "2222222222"
> close(writ)
> unlink("tp.bz2")
c Function
c() function combines its arguments.
c(..., recursive=FALSE)
...: variables to be concatenated
recursive: logical. If recursive = TRUE, the function recursively descends through lists (and pairlists) combining all their elements into a vector
> x <- c(1,2,3,4)
> x
[1] 1 2 3 4
call Function
call() function creates or tests for objects of mode "call".
call(name, ...)
is.call(x)
as.call(x)
name: a non-empty character string naming the function to be called
x: an arbitrary R object
...: arguments to be part of the call
> x <- call("sin",pi)
> x
sin(3.14159265358979)
> eval(x)
[1] 1.224606e-16
capabilities Function
capabilities() function reports on the optional features which have been compiled into this build of R.
> version
_
platform x86_64-w64-mingw32
arch x86_64
os mingw32
system x86_64, mingw32
status
major 3
minor 0.1
year 2013
month 05
day 16
svn rev 62743
language R
version.string R version 3.0.1 (2013-05-16)
nickname Good Sport
> capabilities()
jpeg png tiff tcltk X11 aqua http/ftp sockets
TRUE TRUE TRUE TRUE FALSE FALSE TRUE TRUE
libxml fifo cledit iconv NLS profmem cairo
TRUE FALSE TRUE TRUE TRUE TRUE TRUE
casefold Function
casefold() function translates characters in character vectors, in particular from upper to lower case or vice versa.
casefold(x, upper=FALSE)
x: character vector
...
> x <- "Endmemo"
> x
[1] "Endmemo"
> casefold(x)
[1] "endmemo"
> casefold(x, upper=TRUE)
[1] "ENDMEMO"
cat Function
cat() function prints the objects, concatenates the representations.
cat(... , file = "", sep = " ", fill = FALSE, labels = NULL,
append = FALSE)
...: object
file: print to file
> x <- "r tutorial\n"
> cat(x)
r tutorial
cbind Function
cbind() function combines vector, matrix or data frame by columns.
cbind(x1,x2,...)
x1,x2:vector, matrix, data frames
data1.csv:
data2.csv:
Read in the data from the file:
>x <- read.csv("data1.csv",header=T,sep=",")
>x2 <- read.csv("data2.csv",header=T,sep=",")
>x3 <- cbind(x,x2)
>x3
Subtype Gender Expression Age City
1 A m -0.54 32 New York
2 A f -0.80 21 Houston
3 B f -1.03 34 Seattle
4 C m -0.41 67 Houston
The row number of the two datasets must be equal.
ceiling Function
ceiling() function returns the smallest integers
larger than the parameter.
ceiling(x)
x: numeric variable or vector
> x <- 2.5
> ceiling(x)
[1] 3
> x <- c(3.5, 2.67, 6.2)
> ceiling(x)
[1] 4 3 7
char.expand Function
char.expand() function Seeks a unique match of its first argument among the elements of its second. If successful, it returns this element; otherwise, it performs an action specified by the third argument.
char.expand(input, target, nomatch = stop("no match"))
input: character string to be expanded
target: character vector with the values to be matched against
nomatch: an R expression to be evaluated in case expansion was not possible
> x <- c("sand","and","land")
> char.expand("an",x,warning("no expand"))
[1] "and"
> char.expand("a",x,warning("no expand"))
[1] "and"
> char.expand("xx",x,warning("no expand"))
[1] NA
Warning message:
In eval(nomatch) : no expand
character Function
character() function creates or test for character objects.
character(length = 0)
as.character(x, ...)
is.character(x)
length: A non-negative integer specifying the desired length. Double values will be coerced to integer: supplying an argument of length other than one will give a warning
x: object for test
...
> x <- character()
> x
character(0)
> x <- character(length=5)
> x
[1] "" "" "" "" ""
> x <- 4 + 5
> x
[1] 9
> is.character(x)
[1] FALSE
> as.character(x)
[1] "9"
> y <- as.character(x)
> is.character(y)
[1] TRUE
charmatch Function
charmatch() function finds matches between two arguments.
charmatch(x, table, nomatch = NA_integer_)
x: the values to be matched
table: the values to be matched against
nomatch: the (integer) value to be returned at non-matching positions
...
> charmatch("an",c("and","sand"))
[1] 1
> charmatch("an",c("end","and","sand"))
[1] 2
> charmatch("an","sand")
[1] NA
charToRaw Function
charToRaw() function converts character to ASCII or "raw" objects.
charToRaw(x)
x: character to be converted
...
> x <- "endmemo r tutorial"
> y <- charToRaw(x)
> y
[1] 65 6e 64 6d 65 6d 6f 20 72 20 74 75 74 6f 72 69 61 6c
> x <- charToRaw("a")
> x
[1] 61
chartr Function
chartr() function do string substitutions.
chartr(old, new, x)
old: old string to be substituted
new: new string
x: target string
> x <- "endmemo r tutorial"
> chartr("mdi","gfo",x)
[1] "enfgego r tutoroal"
Chi Square Test Example
chisq.test() function performs chi squared contingency table tests and goodness of fit tests.
chisq.test(x, y = NULL, correct = TRUE, p = rep(1/length(x), length(x)), rescale.p = FALSE, simulate.p.value = FALSE, B = 2000)
•
x: a numeric vector or matrix.
•
y: a numeric vector or a factor (if x is a factor of same length) or NULL (if x is a matrix).
•
correct: a logical indicating whether to apply continuity correction when computing the test statistic for 2 by 2 tables:
one half is subtracted from all |O - E| differences. No correction is done if simulate.p.value = TRUE.
•
p: a vector of probabilities of the same length of x. An error is given if any entry of p is negative.
•
rescale.p: a logical scalar; if TRUE then p is rescaled (if necessary) to sum to 1. If rescale.p is FALSE, and p does not sum to 1, an error is given.
•
simulate.p.value: a logical indicating whether to compute p-values by Monte Carlo simulation.
•
B: an integer specifying the number of replicates used in the Monte Carlo test.
For Example, there are 205 mutations in gene p53 of 514 tumors, while 96 stage IV tumors have 86 mutations.
We expect that 96 stage IV tumors should have 96 x 205 / 514 = 38 mutations, while we observed 86.
Is that significantly different from the general mutation pattern?
The R source code for a chi square goodness of fit test is:
> sam <- matrix(c(86,96,38,96),nrow=2,ncol=2)
> sam
[,1] [,2]
[1,] 86 38
[2,] 96 96
> chisq.test(sam)
Pearson's Chi-squared test with Yates' continuity correction
data: sam
X-squared = 10.7773, df = 1, p-value = 0.001028
> chisq.test(sam)$p.value
[1] 0.001027552
Following is a csv file example.
Following R code can do chi square test of every line in the example file:
x<-read.csv("chisq.csv",header=T,sep=",",dec=".")
zz <- file("out_chisq.txt","w")
title <- names(x)
writeLines(paste(title[1],title[2],title[3],title[4],title[5],
"Chisq P Value",sep=","),con=zz,sep="\n")
xR <- nrow(x)
sam<-array(dim=c(2,2))
for (i in 1:xR)
{
sam[1,] <- c(x[i,2],x[i,3])
sam[2,] <- c(x[i,4],x[i,5])
pv<- chisq.test(sam)$p.value
writeLines(paste(x[i,1],x[i,2],x[i,3],x[i,4],x[i,5],pv,sep=","),
con=zz,sep="\n")
}
close(zz)
The content of the output file is:
Gene,Unique.observed,Unique.expected,duplicated.observed,
duplicate.expected,Chisq P Value
TTN,27,33,60,54,0.425175749168081
GATA3,38,20,17,35,0.00116789922038592
HLA-DRB6,18,15,24,27,0.655008761576397
MUC16,13,15,28,26,0.815855072976336
NR1H2,11,15,29,25,0.473920420172139
GPRIN2,12,14,27,25,0.810181236410474
MAP3K1,15,14,24,25,1
GPRIN1,13,14,25,24,1
MLL3,12,14,26,24,0.808944275014528
MAP3K4,8,14,29,23,0.203492032204285
CDH1,17,12,17,22,0.326688384050414
ENSG00000245549,15,12,18,21,0.616574005797083
ZNF384,12,12,20,20,0.796253414737639
FRG1B,11,11,20,20,0.790676108831151
AKD1,9,11,21,19,0.784191229401619
OBSCN,12,11,17,18,1
NCOA3,8,10,20,18,0.77477725929156
USH2A,8,10,20,18,0.77477725929156
ENSG00000198786,12,10,15,17,0.781814003488769
Download the csv file and the R source code:
Data File
R Source Code File
chol Function
chol() function compute the Choleski factorization of a real symmetric positive-definite
square matrix.
chol(x, ...)
x: an object for which a method exists. The default method applies to real symmetric, positive-definite matrices
...
> x <- matrix(c(8,1,1,4),2,2)
> x
[,1] [,2]
[1,] 8 1
[2,] 1 4
> y <- chol(x)
> y
[,1] [,2]
[1,] 2.828427 0.3535534
[2,] 0.000000 1.9685020
> x <- matrix(rep(1:4),2,2)
> x
[,1] [,2]
[1,] 1 3
[2,] 2 4
> y <- chol(x)
Error in chol.default(x) :
the leading minor of order 2 is not positive definite
chol2inv Function
chol2inv() function inverts a symmetric, positive definite square matrix from its Choleski decomposition.
chol2inv(x, size = NCOL(x), LINPACK = FALSE)
x: matrix
size: the number of columns of x containing the Choleski decomposition
LINPACK: logical. Should LINPACK be used (for compatibility with R < 1.7.0)
> x <- matrix(c(8,1,2,4),2,2)
> x
[,1] [,2]
[1,] 8 2
[2,] 1 4
> y <- chol2inv(x)
> y
[,1] [,2]
[1,] 0.01953125 -0.015625
[2,] -0.01562500 0.062500
choose Function
choose() function computes the combination nCr.
choose(n,r)
n: n elements
r: r subset elements
...
nCr = n!/(r! * (n-r)!)
> choose(5,2)
[1] 10
> choose(2,1)
[1] 2
Draw Circle
draw.circle(...) function draws a circle on the plot. It's usage is:
draw.circle(x,y,radius,nv=100,border=NULL,col=NA,lty=1,lwd=1)
x,y: Circle center coordinates
radius: Circle radius
nv: Number of vertices
border: Border Color
col: Fill Color
lty: Line type
lwd: Line width
draw.circle requires "plotrix" package, to install:
>install.packages("plotrix")
Let's first plot the BOD data frame:
>plot(BOD)

Add a circle to the plot:
>require(plotrix)
>draw.circle(4,14,2,border="blue",col="tan2")

Object Classes
R possesses a simple generic function mechanism which can be used for an object-oriented style of programming. Method dispatch takes place based on the class of the first argument to the generic function.
class(x)
class(x) <- value
unclass(x)
inherits(x, what, which = FALSE)
oldClass(x)
oldClass(x) <- value
x: R object
what, value: character vector naming classes
which: logical affecting return
> x <- c(3,5)
> class(x)
[1] "numeric"
> oldClass(x)
NULL
> inherits(x,c("numeric"))
[1] TRUE
> inherits(x,c("character"))
[1] FALSE
clipboard Function
readClipboard() function reads in from the clipboard.
close Function
close() function close an open handle.
close(handle, type = "rw", ...)
handle: an open file handle
...
> handle <- open(handle, open="r")
> close(handle)
Clustering Tree Plot
Let's first have a look of our data file named clustering.csv:
elements S1 S2 S3 S4 S5 S6 S7 S8
R1 -0.0027 0.1057 0.1976 0.0209 0 0.0089 0.0082 0.0209
R2 0 -0.1204 0.2627 0 0 0.283 0.2076 -0.0158
R3 0 -0.1204 0.2627 0 0 0.283 0.2076 -0.0158
R4 0.0142 0 -0.454 0.0101 -0.0213 -0.0084 -0.0121 0.0083
R5 0 0 -0.2334 0.007 0.4151 0 0.0987 0.021
R6 0.0381 0.0644 0.2302 0 0 -0.0476 0.2432 -0.0069
R7 0.0381 0.0644 0.2302 0 0 -0.0476 0.2432 -0.0069
R8 0.0381 0.0644 0.2302 0 0 -0.0476 0.2432 -0.0069
R9 0.0891 -0.1022 -0.4466 -0.4877 -0.0175 -0.0523 -0.4792 -0.0547
R10 0.0046 -0.1539 -0.4645 0 -0.0282 0 -0.0217 0.017
R11 0.0706 0.028 0.3626 0 0.0196 -0.0094 0.3086 0
R12 0.0311 0.0759 0.2119 0 -0.0022 0 0 0.0117
R13 0.0013 0.0702 -0.3176 0.0152 0.0095 -0.0224 0.2069 0.005
R14 0.0491 0.0525 -0.4329 0.0237 -0.0038 -0.0224 0.2065 0.005
R15 0.0256 0.0579 0.1846 0.0024 0.0029 -0.0165 0.4781 -0.0123
R16 -0.0061 -0.1554 -0.0635 0.0121 -0.0282 0 -0.016 0.017
R17 -0.0061 -0.1554 -0.0635 0.0121 -0.0282 0 -0.016 0.017
A simple unsupervised hierarchical clustering:
>x <- read.csv("clustering.csv", header=T, dec=".",sep=",")
>data.hclust <- hclust(dist(t(x[,2:ncol(x)])),method="complete")
>plot(data.hclust)

Let's add some annotations:
>label <- data.hclust$labels
>for (i in 1:length(label)){
> if (i %% 2 == 1) {label[i]<- paste("control_",label[i],sep="");}
>}
>data.hclust$labels <- label
>plot(data.hclust,pointsize=15,units="px",
+ main="Hierarchical Clustering",xlab="Samples")
>rect.hclust(data.hclust,k=4,border="blue")
>groups<-cutree(data.hclust,k=4)

coef Function
coef() function extracts model coefficients from objects returned by modeling functions.
It's an alias of coefficients().
>x <- c(2,1,3,2,5,3.3,1);
>y <- c(4,2,6,3,8,6,2.2);
Plot the data:

Calculate the coefficients of linear model:
>m < lm(y~x) #Linear Regression Model
>c <- coef(lm(y~x))
>c
(Intercept) x
0.5487805 1.5975610
Draw the regression line:
>abline(c, col="blue")

Calculate the Correlation Coefficient (r
2):
>cr = cor(y,x,method="pearson")
>cr = round(cr,digits=3)
>cr
[1] 0.978
col Function
col() function gets the column number of a matrix.
col(x, as.factor=FALSE)
x: matrix
as.factor: a logical value indicating whether the value should be returned as a factor of column labels (created if necessary) rather than as numbers
...
> x <- matrix(rep(1:9),3,3)
> x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> col(x)
[,1] [,2] [,3]
[1,] 1 2 3
[2,] 1 2 3
[3,] 1 2 3
colMeans Function
colMeans() function computes the means of columns of matrix.
colMeans(x, na.rm = FALSE, dims = 1)
x: array of two or more dimensions, containing numeric, complex, integer or logical values, or a numeric data frame
...
> x <- matrix(rep(1:9),3,3)
> x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> colMeans(x)
[1] 2 5 8
colnames Function
colnames() function retrieve or set the column names of matrix.
colnames(x, do.NULL = TRUE, prefix = "col")
colnames(x) <- value
x: matrix
do.NULL: logical. Should this create names if they are NULL?
prefix: for created names
value: a valid value for that component of dimnames(x)
Following is a csv file example:
Let first read in the data from the file:
> x <- read.csv("matrix.csv",header=T,sep="\t")
> colnames(x)
[1] "A1" "A2" "B1" "B2" "C1" "C2"
> x <- as.matrix(BOD)
> x
Time demand
[1,] 1 8.3
[2,] 2 10.3
[3,] 3 19.0
[4,] 4 16.0
[5,] 5 15.6
[6,] 7 19.8
> is.matrix(x)
[1] TRUE
> colnames(x)
[1] "Time" "demand"
Change the column names:
> colnames(x) <- c("No.","Value")
> x
No. Value
[1,] 1 8.3
[2,] 2 10.3
[3,] 3 19.0
[4,] 4 16.0
[5,] 5 15.6
[6,] 7 19.8
Colors Chart
R has 657 built-in color names. The function colors() will show all of them.
All these color names can be used in plot parameters like col=. The function
col2rgb() can convert all these colors into RGB numbers.
| white |
aliceblue |
antiquewhite |
antiquewhite1 |
| antiquewhite2 |
antiquewhite3 |
antiquewhite4 |
aquamarine |
| aquamarine1 |
aquamarine2 |
aquamarine3 |
aquamarine4 |
| azure |
azure1 |
azure2 |
azure3 |
| azure4 |
beige |
bisque |
bisque1 |
| bisque2 |
bisque3 |
bisque4 |
black |
| blanchedalmond |
blue |
blue1 |
blue2 |
| blue3 |
blue4 |
blueviolet |
brown |
| brown1 |
brown2 |
brown3 |
brown4 |
| burlywood |
burlywood1 |
burlywood2 |
burlywood3 |
| burlywood4 |
cadetblue |
cadetblue1 |
cadetblue2 |
| cadetblue3 |
cadetblue4 |
chartreuse |
chartreuse1 |
| chartreuse2 |
chartreuse3 |
chartreuse4 |
chocolate |
| chocolate1 |
chocolate2 |
chocolate3 |
chocolate4 |
| coral |
coral1 |
coral2 |
coral3 |
| coral4 |
cornflowerblue |
cornsilk |
cornsilk1 |
| cornsilk2 |
cornsilk3 |
cornsilk4 |
cyan |
| cyan1 |
cyan2 |
cyan3 |
cyan4 |
| darkblue |
darkcyan |
darkgoldenrod |
darkgoldenrod1 |
| darkgoldenrod2 |
darkgoldenrod3 |
darkgoldenrod4 |
darkgray |
| darkgreen |
darkgrey |
darkkhaki |
darkmagenta |
| darkolivegreen |
darkolivegreen1 |
darkolivegreen2 |
darkolivegreen3 |
| darkolivegreen4 |
darkorange |
darkorange1 |
darkorange2 |
| darkorange3 |
darkorange4 |
darkorchid |
darkorchid1 |
| darkorchid2 |
darkorchid3 |
darkorchid4 |
darkred |
| darksalmon |
darkseagreen |
darkseagreen1 |
darkseagreen2 |
| darkseagreen3 |
darkseagreen4 |
darkslateblue |
darkslategray |
| darkslategray1 |
darkslategray2 |
darkslategray3 |
darkslategray4 |
| darkslategrey |
darkturquoise |
darkviolet |
deeppink |
| deeppink1 |
deeppink2 |
deeppink3 |
deeppink4 |
| deepskyblue |
deepskyblue1 |
deepskyblue2 |
deepskyblue3 |
| deepskyblue4 |
dimgray |
dimgrey |
dodgerblue |
| dodgerblue1 |
dodgerblue2 |
dodgerblue3 |
dodgerblue4 |
| firebrick |
firebrick1 |
firebrick2 |
firebrick3 |
| firebrick4 |
floralwhite |
forestgreen |
gainsboro |
| ghostwhite |
gold |
gold1 |
gold2 |
| gold3 |
gold4 |
goldenrod |
goldenrod1 |
| goldenrod2 |
goldenrod3 |
goldenrod4 |
gray |
| gray0 |
gray1 |
gray2 |
gray3 |
| gray4 |
gray5 |
gray6 |
gray7 |
| gray8 |
gray9 |
gray10 |
gray11 |
| gray12 |
gray13 |
gray14 |
gray15 |
| gray16 |
gray17 |
gray18 |
gray19 |
| gray20 |
gray21 |
gray22 |
gray23 |
| gray24 |
gray25 |
gray26 |
gray27 |
| gray28 |
gray29 |
gray30 |
gray31 |
| gray32 |
gray33 |
gray34 |
gray35 |
| gray36 |
gray37 |
gray38 |
gray39 |
| gray40 |
gray41 |
gray42 |
gray43 |
| gray44 |
gray45 |
gray46 |
gray47 |
| gray48 |
gray49 |
gray50 |
gray51 |
| gray52 |
gray53 |
gray54 |
gray55 |
| gray56 |
gray57 |
gray58 |
gray59 |
| gray60 |
gray61 |
gray62 |
gray63 |
| gray64 |
gray65 |
gray66 |
gray67 |
| gray68 |
gray69 |
gray70 |
gray71 |
| gray72 |
gray73 |
gray74 |
gray75 |
| gray76 |
gray77 |
gray78 |
gray79 |
| gray80 |
gray81 |
gray82 |
gray83 |
| gray84 |
gray85 |
gray86 |
gray87 |
| gray88 |
gray89 |
gray90 |
gray91 |
| gray92 |
gray93 |
gray94 |
gray95 |
| gray96 |
gray97 |
gray98 |
gray99 |
| gray100 |
green |
green1 |
green2 |
| green3 |
green4 |
greenyellow |
grey |
| grey0 |
grey1 |
grey2 |
grey3 |
| grey4 |
grey5 |
grey6 |
grey7 |
| grey8 |
grey9 |
grey10 |
grey11 |
| grey12 |
grey13 |
grey14 |
grey15 |
| grey16 |
grey17 |
grey18 |
grey19 |
| grey20 |
grey21 |
grey22 |
grey23 |
| grey24 |
grey25 |
grey26 |
grey27 |
| grey28 |
grey29 |
grey30 |
grey31 |
| grey32 |
grey33 |
grey34 |
grey35 |
| grey36 |
grey37 |
grey38 |
grey39 |
| grey40 |
grey41 |
grey42 |
grey43 |
| grey44 |
grey45 |
grey46 |
grey47 |
| grey48 |
grey49 |
grey50 |
grey51 |
| grey52 |
grey53 |
grey54 |
grey55 |
| grey56 |
grey57 |
grey58 |
grey59 |
| grey60 |
grey61 |
grey62 |
grey63 |
| grey64 |
grey65 |
grey66 |
grey67 |
| grey68 |
grey69 |
grey70 |
grey71 |
| grey72 |
grey73 |
grey74 |
grey75 |
| grey76 |
grey77 |
grey78 |
grey79 |
| grey80 |
grey81 |
grey82 |
grey83 |
| grey84 |
grey85 |
grey86 |
grey87 |
| grey88 |
grey89 |
grey90 |
grey91 |
| grey92 |
grey93 |
grey94 |
grey95 |
| grey96 |
grey97 |
grey98 |
grey99 |
| grey100 |
honeydew |
honeydew1 |
honeydew2 |
| honeydew3 |
honeydew4 |
hotpink |
hotpink1 |
| hotpink2 |
hotpink3 |
hotpink4 |
indianred |
| indianred1 |
indianred2 |
indianred3 |
indianred4 |
| ivory |
ivory1 |
ivory2 |
ivory3 |
| ivory4 |
khaki |
khaki1 |
khaki2 |
| khaki3 |
khaki4 |
lavender |
lavenderblush |
| lavenderblush1 |
lavenderblush2 |
lavenderblush3 |
lavenderblush4 |
| lawngreen |
lemonchiffon |
lemonchiffon1 |
lemonchiffon2 |
| lemonchiffon3 |
lemonchiffon4 |
lightblue |
lightblue1 |
| lightblue2 |
lightblue3 |
lightblue4 |
lightcoral |
| lightcyan |
lightcyan1 |
lightcyan2 |
lightcyan3 |
| lightcyan4 |
lightgoldenrod |
lightgoldenrod1 |
lightgoldenrod2 |
| lightgoldenrod3 |
lightgoldenrod4 |
lightgoldenrodyellow |
lightgray |
| lightgreen |
lightgrey |
lightpink |
lightpink1 |
| lightpink2 |
lightpink3 |
lightpink4 |
lightsalmon |
| lightsalmon1 |
lightsalmon2 |
lightsalmon3 |
lightsalmon4 |
| lightseagreen |
lightskyblue |
lightskyblue1 |
lightskyblue2 |
| lightskyblue3 |
lightskyblue4 |
lightslateblue |
lightslategray |
| lightslategrey |
lightsteelblue |
lightsteelblue1 |
lightsteelblue2 |
| lightsteelblue3 |
lightsteelblue4 |
lightyellow |
lightyellow1 |
| lightyellow2 |
lightyellow3 |
lightyellow4 |
limegreen |
| linen |
magenta |
magenta1 |
magenta2 |
| magenta3 |
magenta4 |
maroon |
maroon1 |
| maroon2 |
maroon3 |
maroon4 |
mediumaquamarine |
| mediumblue |
mediumorchid |
mediumorchid1 |
mediumorchid2 |
| mediumorchid3 |
mediumorchid4 |
mediumpurple |
mediumpurple1 |
| mediumpurple2 |
mediumpurple3 |
mediumpurple4 |
mediumseagreen |
| mediumslateblue |
mediumspringgreen |
mediumturquoise |
mediumvioletred |
| midnightblue |
mintcream |
mistyrose |
mistyrose1 |
| mistyrose2 |
mistyrose3 |
mistyrose4 |
moccasin |
| navajowhite |
navajowhite1 |
navajowhite2 |
navajowhite3 |
| navajowhite4 |
navy |
navyblue |
oldlace |
| olivedrab |
olivedrab1 |
olivedrab2 |
olivedrab3 |
| olivedrab4 |
orange |
orange1 |
orange2 |
| orange3 |
orange4 |
orangered |
orangered1 |
| orangered2 |
orangered3 |
orangered4 |
orchid |
| orchid1 |
orchid2 |
orchid3 |
orchid4 |
| palegoldenrod |
palegreen |
palegreen1 |
palegreen2 |
| palegreen3 |
palegreen4 |
paleturquoise |
paleturquoise1 |
| paleturquoise2 |
paleturquoise3 |
paleturquoise4 |
palevioletred |
| palevioletred1 |
palevioletred2 |
palevioletred3 |
palevioletred4 |
| papayawhip |
peachpuff |
peachpuff1 |
peachpuff2 |
| peachpuff3 |
peachpuff4 |
peru |
pink |
| pink1 |
pink2 |
pink3 |
pink4 |
| plum |
plum1 |
plum2 |
plum3 |
| plum4 |
powderblue |
purple |
purple1 |
| purple2 |
purple3 |
purple4 |
red |
| red1 |
red2 |
red3 |
red4 |
| rosybrown |
rosybrown1 |
rosybrown2 |
rosybrown3 |
| rosybrown4 |
royalblue |
royalblue1 |
royalblue2 |
| royalblue3 |
royalblue4 |
saddlebrown |
salmon |
| salmon1 |
salmon2 |
salmon3 |
salmon4 |
| sandybrown |
seagreen |
seagreen1 |
seagreen2 |
| seagreen3 |
seagreen4 |
seashell |
seashell1 |
| seashell2 |
seashell3 |
seashell4 |
sienna |
| sienna1 |
sienna2 |
sienna3 |
sienna4 |
| skyblue |
skyblue1 |
skyblue2 |
skyblue3 |
| skyblue4 |
slateblue |
slateblue1 |
slateblue2 |
| slateblue3 |
slateblue4 |
slategray |
slategray1 |
| slategray2 |
slategray3 |
slategray4 |
slategrey |
| snow |
snow1 |
snow2 |
snow3 |
| snow4 |
springgreen |
springgreen1 |
springgreen2 |
| springgreen3 |
springgreen4 |
steelblue |
steelblue1 |
| steelblue2 |
steelblue3 |
steelblue4 |
tan |
| tan1 |
tan2 |
tan3 |
tan4 |
| thistle |
thistle1 |
thistle2 |
thistle3 |
| thistle4 |
tomato |
tomato1 |
tomato2 |
| tomato3 |
tomato4 |
turquoise |
turquoise1 |
| turquoise2 |
turquoise3 |
turquoise4 |
violet |
| violetred |
violetred1 |
violetred2 |
violetred3 |
| violetred4 |
wheat |
wheat1 |
wheat2 |
| wheat3 |
wheat4 |
whitesmoke |
yellow |
| yellow1 |
yellow2 |
yellow3 |
yellow4 |
| yellowgreen |
|
colSums Function
colSums() function computes the sums of matrix columns.
colSums (x, na.rm = FALSE, dims = 1)
x: matrix
...
> x <- matrix(rep(1:9),3,3)
> x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> colSums(x)
[1] 6 15 24
commandArgs Function
commandArgs() function prints the command line arguments.
> commandArgs()
[1] "C:\\Program Files\\R\\R-3.0.1\\bin\\x64\\Rgui.exe"
comment Function
comment() function sets or queries a comment attribute for an objects.
comment(x)
comment(x) <- value
x: an object
value: comment string
> x <- 3.1415926
> comment(x) <- "pi"
> x
[1] 3.141593
> comment(x)
[1] "pi"
complex Function
complex() function do complex number calculations.
complex(length.out = 0, real = numeric(), imaginary = numeric(),
modulus = 1, argument = 0)
as.complex(x, ...)
is.complex(x)
Re(z)
Im(z)
Mod(z)
Arg(z)
Conj(z)
length.out: numeric. Desired length of the output vector, inputs being recycled as needed
real: numeric vector
imaginary: numeric vector
modulus: numberic vector
argument:
x, z: complex object
...
require(graphics)
0i ^ (-3:3)
matrix(1i^ (-6:5), nrow=4) #- all columns are the same
0 ^ 1i # a complex NaN
## create a complex normal vector
z <- complex(real = stats::rnorm(100), imaginary = stats::rnorm(100))
## or also (less efficiently):
z2 <- 1:2 + 1i*(8:9)
## The Arg(.) is an angle:
zz <- (rep(1:4,len=9) + 1i*(9:1))/10
zz.shift <- complex(modulus = Mod(zz), argument= Arg(zz) + pi)
plot(zz, xlim=c(-1,1), ylim=c(-1,1), col="red", asp = 1,
main = expression(paste("Rotation by "," ", pi == 180^o)))
abline(h=0,v=0, col="blue", lty=3)
points(zz.shift, col="orange")
Compress and Decompress
memCompress() and memDecompress() functions conducts in-memory
compression or decompression for raw vectors.
memCompress(from, type = c("gzip", "bzip2", "xz", "none"))
memDecompress(from,
type = c("unknown", "gzip", "bzip2", "xz", "none"),
asChar = FALSE)
from: raw vector
type: type of compression
asChar: whether convert the result to character string or not
...
> txt <- readLines(file.path(R.home("doc"), "COPYING"))
> sum(nchar(txt))
[1] 17671
> txt.gz <- memCompress(txt,"g")
> length(txt.gz)
[1] 6837
condition handling
R has a series of functions to handle unusual conditions, including errors and warnings.
tryCatch(expr, ..., finally)
withCallingHandlers(expr, ...)
signalCondition(cond)
simpleCondition(message, call = NULL)
simpleError (message, call = NULL)
simpleWarning (message, call = NULL)
simpleMessage (message, call = NULL)
## S3 method for class 'condition'
as.character(x, ...)
## S3 method for class 'error'
as.character(x, ...)
## S3 method for class 'condition'
print(x, ...)
## S3 method for class 'restart'
print(x, ...)
conditionCall(c)
## S3 method for class 'condition'
conditionCall(c)
conditionMessage(c)
## S3 method for class 'condition'
conditionMessage(c)
withRestarts(expr, ...)
computeRestarts(cond = NULL)
findRestart(name, cond = NULL)
invokeRestart(r, ...)
invokeRestartInteractively(r)
isRestart(x)
restartDescription(r)
restartFormals(r)
.signalSimpleWarning(msg, call)
.handleSimpleError(h, msg, call)
c: condition object
call: call expression
cond: a condition object
expr: expression to be evaluated
finally: expression to be evaluated before returning or exiting
h: function
r: restart object
...
tryCatch(1, finally=print("Hello"))
e <- simpleError("test error")
## Not run:
stop(e)
tryCatch(stop(e), finally=print("Hello"))
tryCatch(stop("fred"), finally=print("Hello"))
## End(Not run)
tryCatch(stop(e), error = function(e) e, finally=print("Hello"))
tryCatch(stop("fred"), error = function(e) e, finally=print("Hello"))
withCallingHandlers({ warning("A"); 1+2 }, warning = function(w) {})
## Not run:
{ withRestarts(stop("A"), abort = function() {}); 1 }
## End(Not run)
withRestarts(invokeRestart("foo", 1, 2), foo = function(x, y) {x + y})
conflicts Function
conflicts() function conflicts reports on objects that exist with the same name in two or more places on the search path, usually because an object in the user's workspace or a package is masking a system object of the same name. This helps discover unintentional masking.
conflicts(where = search(), detail = FALSE)
where: A subset of the search path, by default the whole search path
detail: If TRUE, give the masked or masking functions for all members of the search path.
...
> conflicts()
[1] "body<-" "kronecker"
Connections
showConnections(all = FALSE)
getConnection(what)
closeAllConnections()
stdin()
stdout()
stderr()
isatty(con)
stdin(), stdout() and stderr() are standard connections corresponding to input, output and error on the console respectively (and not necessarily to file streams). They are text-mode connections of class "terminal" which cannot be opened or closed, and are read-only, write-only and write-only respectively. The stdout() and stderr() connections can be re-directed by sink (and in some circumstances the output from stdout() can be split: see the help page).
The encoding for stdin() when redirected can be set by the command-line flag --encoding.
showConnections returns a matrix of information. If a connection object has been lost or forgotten, getConnection will take a row number from the table and return a connection object for that connection, which can be used to close the connection, for example. However, if there is no R level object referring to the connection it will be closed automatically at the next garbage collection.
closeAllConnections closes (and destroys) all user connections, restoring all sink diversions as it does so.
isatty returns true if the connection is one of the class "terminal" connections and it is apparently connected to a terminal, otherwise false. This may not be reliable in embedded applications, including GUI consoles.
Built-in Constants
R built-in Constants includes:
• LETTERS: 26 letters in uppercase
• letters: 26 letters in lowercase
• month.abb: 12 month names in abbreviation form
• month.name: 12 month names in full name
• pi: π
> LETTERS
[1] "A" "B" "C" "D" "E" "F" "G" "H" "I" "J" "K" "L" "M" "N"
[15] "O" "P" "Q" "R" "S" "T" "U" "V" "W" "X" "Y" "Z"
> letters
[1] "a" "b" "c" "d" "e" "f" "g" "h" "i" "j" "k" "l" "m" "n"
[15] "o" "p" "q" "r" "s" "t" "u" "v" "w" "x" "y" "z"
> month.abb
[1] "Jan" "Feb" "Mar" "Apr" "May" "Jun" "Jul"
[8] "Aug" "Sep" "Oct" "Nov" "Dec"
> month.name
[1] "January" "February" "March" "April" "May" "June"
[7] "July" "August" "September" "October" "November" "December"
> pi
[1] 3.141593
contributors Function
contributors() function prints out all the contributors of R development.
> contributors()
R is a project which is attempting to provide a modern piece of
statistical software for the GNU suite of software.
The current R is the result of a collaborative effort with
contributions from all over the world.
Authors of R.
R was initially written by Robert Gentleman and Ross Ihaka of the
Statistics Department of the University of Auckland.
Since mid-1997 there has been a core group with write access to the R
source, currently consisting of
Douglas Bates
John Chambers
Peter Dalgaard
Seth Falcon
Robert Gentleman
Kurt Hornik
Stefano Iacus
Ross Ihaka
Friedrich Leisch
Uwe Ligges
Thomas Lumley
Martin Maechler
Duncan Murdoch
Paul Murrell
Martyn Plummer
Brian Ripley
Deepayan Sarkar
Duncan Temple Lang
Luke Tierney
Simon Urbanek
plus Heiner Schwarte up to October 1999 and Guido Masarotto up to June 2003.
Current R-core members can be contacted via email to R-project.org
with name made up by replacing spaces by dots in the name listed above.
R would not be what it is today without the invaluable help of these
people, who contributed by donating code, bug fixes and documentation:
Valerio Aimale, Thomas Baier, Henrik Bengtsson, Roger Bivand,
Ben Bolker, David Brahm, Goran Brostrom, Patrick Burns, Vince Carey,
Saikat DebRoy, Brian D'Urso, Lyndon Drake, Dirk Eddelbuettel,
Claus Ekstrom, Sebastian Fischmeister, John Fox, Paul Gilbert,
Yu Gong, Gabor Grothendieck, Frank E Harrell Jr, Torsten Hothorn,
Robert King, Kjetil Kjernsmo, Roger Koenker, Philippe Lambert,
Jan de Leeuw, Jim Lindsey, Patrick Lindsey, Catherine Loader,
Gordon Maclean, John Maindonald, David Meyer, Ei-ji Nakama,
Jens Oehlschaegel, Steve Oncley, Richard O'Keefe, Hubert Palme,
Roger D. Peng, Jose' C. Pinheiro, Tony Plate, Anthony Rossini,
Jonathan Rougier, Petr Savicky, Guenther Sawitzki, Marc Schwartz,
Detlef Steuer, Bill Simpson, Gordon Smyth, Adrian Trapletti,
Terry Therneau, Rolf Turner, Bill Venables, Gregory R. Warnes,
Andreas Weingessel, Morten Welinder, James Wettenhall, Simon Wood and
Achim Zeileis.
Others have written code that has been adopted by R and is
acknowledged in the code files, including
J. D. Beasley, David J. Best, Richard Brent, Kevin Buhr, Michael
A. Covington, Bill Cleveland, Robert Cleveland,, G. W. Cran,
C. G. Ding, Ulrich Drepper, Paul Eggert, J. O. Evans, David M. Gay,
H. Frick, G. W. Hill, Richard H. Jones, Eric Grosse, Shelby Haberman,
Bruno Haible, John Hartigan, Andrew Harvey, Trevor Hastie, Min Long
Lam, George Marsaglia, K. J. Martin, Gordon Matzigkeit,
C. R. Mckenzie, Jean McRae, Cyrus Mehta, Fionn Murtagh, John C. Nash,
Finbarr O'Sullivan, R. E. Odeh, William Patefield, Nitin Patel, Alan
Richardson, D. E. Roberts, Patrick Royston, Russell Lenth, Ming-Jen
Shyu, Richard C. Singleton, S. G. Springer, Supoj Sutanthavibul, Irma
Terpenning, G. E. Thomas, Rob Tibshirani, Wai Wan Tsang, Berwin
Turlach, Gary V. Vaughan, Michael Wichura, Jingbo Wang, M. A. Wong,
and the Free Software Foundation (for autoconf code and utilities).
See also files under src/extras.
Many more, too numerous to mention here, have contributed by sending bug
reports and suggesting various improvements.
Simon Davies whilst at the University of Auckland wrote the original
version of glm().
Julian Harris and Wing Kwong (Tiki) Wan whilst at the University of
Auckland assisted Ross Ihaka with the original Macintosh port.
R was inspired by the S environment which has been principally
developed by John Chambers, with substantial input from Douglas Bates,
Rick Becker, Bill Cleveland, Trevor Hastie, Daryl Pregibon and
Allan Wilks.
A special debt is owed to John Chambers who has graciously contributed
advice and encouragement in the early days of R and later became a
member of the core team.
The R Foundation may decide to give out @R-project.org
email addresses to contributors to the R Project (even without making them
members of the R Foundation) when in the view of the R Foundation this
would help advance the R project.
The R Core Group, Roger Bivand, John Fox and Bill Venables are the
ordinary members of the R Foundation. In addition, Dirk Eddelbuettel,
Torsten Hothorn, David Meyer, Simon Wood, and Achim Zeileis are also
e-addressable by .@R-project.org.
cos Function
cos() function computes the cosine value of numeric value.
cos(x)
x: Numeric value, array or vector
> cos(pi)
[1] -1
> cos(-pi)
[1] -1
> cos(pi/3)
[1] 0.5
> cos(0)
[1] 1
> x <- c(pi, pi/4, pi/3)
> cos(x)
[1] -1.0000000 0.7071068 0.5000000
X
(deg) |
X
(Rad) |
Y=cosine(X) |
| 180 ̊ |
π |
-1 |
| 150 ̊ |
5π/6 |
-0.866025 |
| 135 ̊ |
3π/4 |
-0.707107 |
| 120 ̊ |
2π/3 |
-0.5 |
| 90 ̊ |
π/2 |
0 |
| 60 ̊ |
π/3 |
0.5 |
| 45 ̊ |
π/4 |
0.707107 |
| 30 ̊ |
π/6 |
0.866025 |
| 0 ̊ |
0 |
1 |
cosh Function
cosh() function computes the hyperbolic cosine of numberic data.
cosh(x)
x: Numeric value, array or vector.
> cosh(1)
[1] 1.543081
> cosh(0.5)
[1] 1.127626
> x <- c(1,0.5)
> cosh(x)
[1] 1.543081 1.127626
crossprod Function
crossprod() function returns matrix cross-product.
crossprod(x, y = NULL)
tcrossprod(x, y = NULL)
x: numeric matrix
y: numeric matrix, if y=NULL, y is the same as x
...
> x <- matrix(1:9,3,3)
> x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> crossprod(x)
[,1] [,2] [,3]
[1,] 14 32 50
[2,] 32 77 122
[3,] 50 122 194
> tcrossprod(x)
[,1] [,2] [,3]
[1,] 66 78 90
[2,] 78 93 108
[3,] 90 108 126
Cstack_info Function
Cstack_info() function Reports information on the C stack size and usage (if available).
> Cstack_info()
size current direction eval_depth
67108864 8168 1 2
cummax Function
cummax() function returns the cumulative maxima.
cummax(x)
x: numeric object
...
> cumsum(2:4)
[1] 2 3 4
> x <- c(3,5,9)
> cummax(x)
[1] 3 5 9
> x <- c(3,5,9,2)
> cummax(x)
[1] 3 5 9 9
cummin Function
cummin() function returns the cumulative minima.
cummin(x)
x: numeric object
...
> cummin(2:4)
[1] 2 2 2
> x <- c(3,5,9)
> cummin(x)
[1] 3 3 3
cumprod Function
cumprod() function returns the cumulative multiplication results.
cumsum(x)
x: numeric or complex object
...
> cumsum(2:4)
[1] 2 6 24
> x <- c(3,5,9)
> cumprod(x)
[1] 3 15 135
cumsum Function
cumsum() function returns the cumulative sums.
cumsum(x)
x: numeric object
...
> cumsum(2:4)
[1] 2 5 9
> x <- c(3,5,9)
> cumsum(x)
[1] 3 8 17
cut Function
cut() function divides a numeric vector into different ranges.
cut(x, breaks, labels = NULL,
include.lowest = FALSE, right = TRUE, dig.lab = 3,
ordered_result = FALSE, ...)
•
x: numeric vector
•
breaks: break points, number or numeric vector.
•
labels: level labels, character vector.
...
> x <- stats::rnorm(100)
> x
[1] -0.154103462 0.271704132 -0.234160855 0.764474679 0.438237645
[6] -0.763854668 1.303402711 0.051660328 1.064258570 0.079144697
[11] -0.704381407 2.239763673 -0.749203152 0.601148921 -0.174814689
[16] 0.100238929 0.670921777 -0.351881772 -1.452691553 0.774250401
[21] 0.985238459 -0.159947063 0.456925349 0.062732203 -0.139094156
[26] -0.021987877 -0.369758710 -0.623015605 0.818971164 1.024360342
[31] -1.180039385 -1.126115746 -1.331609773 0.261068252 0.306040509
[36] 0.186887898 0.039764640 0.618133561 0.808466877 1.530479825
[41] -0.326594787 -0.525549355 -0.038649831 -0.320394434 -0.116615568
[46] -0.928403864 1.284014444 0.559523194 0.511753047 -0.093609863
[51] -1.199423552 -0.358438485 -1.421215594 -0.199430722 -1.285244671
[56] -0.344308069 0.202383513 -1.044830704 0.009940864 -1.083693166
[61] 0.985718206 0.942167477 0.077569581 1.456191918 -1.385394960
[66] -0.174887806 -0.869293103 1.051227075 -0.726361522 0.082628666
[71] 1.275779587 0.258221666 -0.629207453 -0.589352154 -0.818233970
[76] 0.028423636 -0.491220068 0.796916741 -1.407925480 0.765093431
[81] -0.263630781 0.854937357 0.592710059 -0.095388956 -1.064601796
[86] 0.691149856 0.822038961 0.666786287 -1.062610036 -2.833961199
[91] 1.570993774 -0.876630726 -0.343492831 -0.480549452 1.494723381
[96] -2.025528709 0.949853574 -0.917568904 -1.103676434 0.728284402
Divide the data into ranges -5 ~ 5:
> c <- cut(x,breaks=-5:5)
> c
[1] (-1,0] (0,1] (-1,0] (0,1] (0,1] (-1,0] (1,2] (0,1] (1,2]
[10] (0,1] (-1,0] (2,3] (-1,0] (0,1] (-1,0] (0,1] (0,1] (-1,0]
[19] (-2,-1] (0,1] (0,1] (-1,0] (0,1] (0,1] (-1,0] (-1,0] (-1,0]
[28] (-1,0] (0,1] (1,2] (-2,-1] (-2,-1] (-2,-1] (0,1] (0,1] (0,1]
[37] (0,1] (0,1] (0,1] (1,2] (-1,0] (-1,0] (-1,0] (-1,0] (-1,0]
[46] (-1,0] (1,2] (0,1] (0,1] (-1,0] (-2,-1] (-1,0] (-2,-1] (-1,0]
[55] (-2,-1] (-1,0] (0,1] (-2,-1] (0,1] (-2,-1] (0,1] (0,1] (0,1]
[64] (1,2] (-2,-1] (-1,0] (-1,0] (1,2] (-1,0] (0,1] (1,2] (0,1]
[73] (-1,0] (-1,0] (-1,0] (0,1] (-1,0] (0,1] (-2,-1] (0,1] (-1,0]
[82] (0,1] (0,1] (-1,0] (-2,-1] (0,1] (0,1] (0,1] (-2,-1] (-3,-2]
[91] (1,2] (-1,0] (-1,0] (-1,0] (1,2] (-3,-2] (0,1] (-1,0] (-2,-1]
[100] (0,1]
10 Levels: (-5,-4] (-4,-3] (-3,-2] (-2,-1] (-1,0] (0,1] (1,2] (2,3] ... (4,5]
Check the data distribution in different ranges:
> summary(c) #or table(c)
c
(-5,-4] (-4,-3] (-3,-2] (-2,-1] (-1,0] (0,1] (1,2] (2,3] (3,4] (4,5]
0 0 2 14 35 38 10 1 0 0
The numbers are divided into 10 levels, the default step is 1.
Some levels are empty.
Let's try just define the total level number:
> x <- stats::rnorm(100) #random numbers, different every time
> c <- cut(x,breaks=10,dig.lab=2)
> summary(c)
(-2,-1.6] (-1.6,-1.1] (-1.1,-0.69] (-0.69,-0.24] (-0.24,0.21]
5 5 13 20 18
(0.21,0.65] (0.65,1.1] (1.1,1.5] (1.5,2] (2,2.4]
12 14 6 3 4
Label all the levles:
> x <- stats::rnorm(100) #random numbers, different every time
> c <- cut(x,breaks=10,dig.lab=2,labels=1:10)
> summary(c)
1 2 3 4 5 6 7 8 9 10
5 5 13 20 18 12 14 6 3 4
Try again, divide into different ranges (break points):
> x <- stats::rnorm(100) #random numbers, different every time
> c <- cut(x,breaks=c(-2,0,1,2))
> table(c)
c
(-2,0] (0,1] (1,2]
52 32 11
data Function
data.class() function determines the class of an object.
data.class(x)
x: R object
> x <- c(3,5,9)
> data.class(x)
[1] "numeric"
> data.class(letters)
[1] "character"
Data Frame Data Type
R data.frame is a powerful data type, especially when processing table (.csv). It can store the data as row and columns according
to the table. The difference between data frame and matrix is that the column data of matrix are the same, while
the column data of data frame may be of different modes and attributes.
Let's use the R Data Sets BOD (Biochemical Oxygen Demand), which is a data frame:
>x <- BOD
>is.matrix(x)
[1] FALSE
>is.data.frame(x)
[1] TURE
>class(x)
[1] "data.frame"
>x
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
as.data.frame() can coerce a list into a data frame, providing that the components of the list
conforms to the restrictions of a data frame.
Each row of the data frame is a
list or a data frame with one row:
>y <- x[2,]
>is.list(y)
[1] TRUE
>is.data.frame(y)
[1] TRUE
Access the column of the data frame:
>x$Time
[1] 1 2 3 4 5 7
>x$demand
[1] 8.3 10.3 19.0 16.0 15.6 19.8
A convenient way to access the columns of a data frame is using attach(), detach() statement.
e.g. after attach(x), the column x$demand can be accessed by simply typing demand.
>attach(x)
>demand
[1] 8.3 10.3 19.0 16.0 15.6 19.8
In other words, attach() statement makes the components of the data frame visible.
We can do some operations with the variable demand, and the components demand of the data frame will not be changed.
>demand <- demand + 10
>demand
[1] 18.3 20.3 29.0 26.0 25.6 29.8
>x$demand
[1] 8.3 10.3 19.0 16.0 15.6 19.8
Statement detach() is the reverse statement of attach().
>detach(x)
>demand
Error: object 'demand' not found
data.frame is the default data type when you read in a table.
Following is a csv table file dataframe.csv, there are "Expression" value vs Subtype "A", "B" and "C" in column 1 and column 2:
Let's read in the data from the file:
>x <- read.csv("dataframe.csv",header=T,sep="\t")
>is.data.frame(x)
[1] TRUE
Date and Time Functions
R has serveral date and time related functions. date() functions returns a date without time
as character string.
Sys.Date() and Sys.time() returns the system's date and time as a Date and
POSIXlt/POSIXct object respectively.
>date()
[1] "Fri Jan 04 17:38:05 2013"
>Sys.time()
[1] "2013-01-04 17:47:39 EST"
>Sys.Date()
[1] "2013-01-04"
>class(date())
[1] "character"
>class(Sys.Date())
[1] "Date"
>class(Sys.time())
[1] "POSIXct" "POSIXt"
POSIXct contains seconds from 1970.
POSIXlt is a list, contains:
sec, 0-61: seconds
min, 0-59: minutes
hour 0-23: hours
mday 1-31: day of the month
mon 0-11: months after the first of the year
year: years since 1900
wday, 0-6: day of the week
yday, 0-365: day of the year
isdst: Daylight savings time flag
>x <- "19:18:05"
>y <- strptime(x,"%H:%M:%S")
>y
[1] "2013-01-04 19:18:05"
>class(y)
[1] "POSIXlt" "POSIXt"
>y$sec
[1] 5
R date time format:
| %a |
Abbreviated weekday name in the current locale. (Also matches full name on input.) |
| %A |
Full weekday name in the current locale. (Also matches abbreviated name on input.) |
| %b |
Abbreviated month name in the current locale. (Also matches full name on input.) |
| %B |
Full month name in the current locale. (Also matches abbreviated name on input.) |
| %c |
Date and time. Locale-specific on output, "%a %b %e %H:%M:%S %Y" on input. |
| %d |
Day of the month as decimal number (01-31). |
| %H |
Hours as decimal number (00-23). As a special exception times such as 24:00:00 are accepted for input, since ISO 8601 allows these. |
| %I |
Hours as decimal number (01-12). |
| %j |
Day of year as decimal number (001-366). |
| %m |
Month as decimal number (01-12). |
| %M |
Minute as decimal number (00-59). |
| %p |
AM/PM indicator in the locale. Used in conjunction with %I and not with %H. An empty string in some locales. |
| %S |
Second as decimal number (00-61), allowing for up to two leap-seconds (but POSIX-compliant implementations will ignore leap seconds). |
| %U |
Week of the year as decimal number (00-53) using Sunday as the first day 1 of the week (and typically with the first Sunday of the year as day 1 of week 1). The US convention. |
| %w |
Weekday as decimal number (0-6, Sunday is 0). |
| %W |
Week of the year as decimal number (00-53) using Monday as the first day of week (and typically with the first Monday of the year as day 1 of week 1). The UK convention. |
| %x |
Date. Locale-specific on output, "%y/%m/%d" on input. |
| %X |
Time. Locale-specific on output, "%H:%M:%S" on input. |
| %y |
Year without century (00-99). On input, values 00 to 68 are prefixed by 20 and 69 to 99 by 19 - that is the behaviour specified by the 2004 and 2008 POSIX standards, but they do also say "it is expected that in a future version the default century inferred from a 2-digit year will change". |
| %Y |
Year with century. Note that whereas there was no zero in the original Gregorian calendar, ISO 8601:2004 defines it to be valid (interpreted as 1BC): see http://en.wikipedia.org/wiki/0_(year). Note that the standard also says that years before 1582 in its calendar should only be used with agreement of the parties involved. |
| %z |
Signed offset in hours and minutes from UTC, so -0800 is 8 hours behind UTC. |
| %Z |
(output only.) Time zone as a character string (empty if not available).
Where leading zeros are shown they will be used on output but are optional on input.
Note that when %z or %Z is used for output with an object with an assigned timezone an attempt is made to use the values for that timezone, but it is not guaranteed to succeed.
|
debug Function
debug() function sets the debugging flag on a function.
debug(f, text="", condition=NULL)
debugonce(fun, text="", condition=NULL)
undebug(fun)
isdebugged(fun)
f: R function
text: a text string that can be retrieved when the browser is entered
condition: a condition that can be retrieved when the browser is entered
Defunct Function
When a function is removed from R it should be replaced by a function which calls .Defunct.
.Defunct(new, package = NULL, msg)
new: character string: A suggestion for a replacement function
package: character string: The package to be used when suggesting where the defunct function might be listed
msg: character string: A message to be printed, if missing a default message is used
> .Defunct
function (new, package = NULL, msg)
{
if (missing(msg)) {
msg <- gettextf("'%s' is defunct.\n",
as.character(sys.call(sys.parent())[[1L]]))
if (!missing(new))
msg <- c(msg, gettextf("Use '%s' instead.\n", new))
msg <- c(msg, if (!is.null(package)) gettextf("See help(\"Defunct
\") and help(\"%s-defunct\").",
package) else gettext("See help(\"Defunct\")"))
}
else msg <- as.character(msg)
stop(paste(msg, collapse = ""), call. = FALSE, domain = NA)
}
delayedAssign Function
delayedAssign() function delayedAssign creates a promise to evaluate the given expression if its value is requested. This provides direct access to the lazy evaluation mechanism used by R for the evaluation of (interpreted) functions.
delayedAssign(x, value, eval.env = parent.frame(1),
assign.env = parent.frame(1))
x: a variable name (given as a quoted string in the function call)
value: an expression to be assigned to x
eval.env: an environment in which to evaluate value
assign.env: an environment in which to assign x
...
> str <- "R tutorial"
> delayedAssign("x",str)
> str <- "Perl"
> x
[1] "Perl"
When the value of str variable changed, the variable x is assigned the new value. However if x was used before the value change,
the new value will not be assigned.
> str <- "R tutorial"
> delayedAssign("x",str)
> x
[1] "R tutorial"
density Function
density() function computes kernel density estimates.
density(x, bw = "nrd0", adjust = 1,
kernel = c("gaussian", "epanechnikov", "rectangular",
"triangular", "biweight",
"cosine", "optcosine"),
weights = NULL, window = kernel, width,
give.Rkern = FALSE,
n = 512, from, to, cut = 3, na.rm = FALSE, ...)
x: number vector
bw: smoothing bandwidth
...
Let generate 100 numbers randomly:
>x <- stats::rnorm(100)
>x
[1] -0.154103462 0.271704132 -0.234160855 0.764474679 0.438237645
[6] -0.763854668 1.303402711 0.051660328 1.064258570 0.079144697
[11] -0.704381407 2.239763673 -0.749203152 0.601148921 -0.174814689
[16] 0.100238929 0.670921777 -0.351881772 -1.452691553 0.774250401
[21] 0.985238459 -0.159947063 0.456925349 0.062732203 -0.139094156
[26] -0.021987877 -0.369758710 -0.623015605 0.818971164 1.024360342
[31] -1.180039385 -1.126115746 -1.331609773 0.261068252 0.306040509
[36] 0.186887898 0.039764640 0.618133561 0.808466877 1.530479825
[41] -0.326594787 -0.525549355 -0.038649831 -0.320394434 -0.116615568
[46] -0.928403864 1.284014444 0.559523194 0.511753047 -0.093609863
[51] -1.199423552 -0.358438485 -1.421215594 -0.199430722 -1.285244671
[56] -0.344308069 0.202383513 -1.044830704 0.009940864 -1.083693166
[61] 0.985718206 0.942167477 0.077569581 1.456191918 -1.385394960
[66] -0.174887806 -0.869293103 1.051227075 -0.726361522 0.082628666
[71] 1.275779587 0.258221666 -0.629207453 -0.589352154 -0.818233970
[76] 0.028423636 -0.491220068 0.796916741 -1.407925480 0.765093431
[81] -0.263630781 0.854937357 0.592710059 -0.095388956 -1.064601796
[86] 0.691149856 0.822038961 0.666786287 -1.062610036 -2.833961199
[91] 1.570993774 -0.876630726 -0.343492831 -0.480549452 1.494723381
[96] -2.025528709 0.949853574 -0.917568904 -1.103676434 0.728284402
>d <- density(x)
>d
Call:
density.default(x = x)
Data: x (100 obs.); Bandwidth 'bw' = 0.3184
x y
Min. :-3.7891 Min. :0.0001413
1st Qu.:-2.0431 1st Qu.:0.0117442
Median :-0.2971 Median :0.0627054
Mean :-0.2971 Mean :0.1430424
3rd Qu.: 1.4489 3rd Qu.:0.2957362
Max. : 3.1949 Max. :0.4192181
Plot the density:
>plot(density(x),xlim=c(-4,4),col="blueviolet")

deparse Function
deparse() function turns unevaluated expressions into character strings.
deparse(expr, width.cutoff = 60L,
backtick = mode(expr) %in%
c("call", "expression", "(", "function"),
control = c("keepInteger", "showAttributes", "keepNA"),
nlines = -1L)
expr: R expression
with.cutoff: integer in [20, 500] determining the cutoff (in bytes) at which line-breaking is tried
backtick: logical indicating whether symbolic names should be enclosed in backticks if they do not follow the standard syntax
control: character vector of deparsing options
nlines: integer: the maximum number of lines to produce. Negative values indicate no limit
...
> deparse(args(lm))
[1] "function (formula, data, subset, weights, na.action, method = \"qr\", "
[2] " model = TRUE, x = FALSE, y = FALSE, qr = TRUE, singular.ok = TRUE, "
[3] " contrasts = NULL, offset, ...) "
[4] "NULL"
> deparse(args(lm), width=20)
[1] "function (formula, data, " " subset, weights, "
[3] " na.action, method = \"qr\", " " model = TRUE, x = FALSE, "
[5] " y = FALSE, qr = TRUE, " " singular.ok = TRUE, "
[7] " contrasts = NULL, " " offset, ...) "
[9] "NULL"
Deprecated Function
When an object is about removed from R it is first deprecated and should include a call to .Deprecated.
.Deprecated(new, package=NULL, msg)
new: suggestion for a replacement function
package: The package to be used when suggesting where the deprecated function might be listed
msg: message to be printed, if missing a default message is used
> .Deprecated
function (new, package = NULL, msg,
old = as.character(sys.call(sys.parent()))[1L])
{
msg <- if (missing(msg)) {
msg <- gettextf("'%s' is deprecated.\n", old)
if (!missing(new))
msg <- c(msg, gettextf("Use '%s' instead.\n", new))
c(msg, if (!is.null(package)) gettextf("See help(\"Deprecated\")
and help(\"%s-deprecated\").",
package) else gettext("See help(\"Deprecated\")"))
}
else as.character(msg)
warning(paste(msg, collapse = ""), call. = FALSE, domain = NA)
}
det Function
det() function calculates the determinant of a matrix. determinant is a generic function that returns separately the modulus of the determinant, optionally on the logarithm scale, and the sign of the determinant.
det(x, ...)
determinant(x, logarithm = TRUE, ...)
x: matrix
logarithm: logical; if TRUE (default) return the logarithm of the modulus of the determinant
...
> x <- matrix(c(-2,2,-3,-1,1,3,2,0,-1),3,3)
> x
[,1] [,2] [,3]
[1,] -2 -1 2
[2,] 2 1 0
[3,] -3 3 -1
> det(x)
[1] 18
> x <- t(x)
> x
[,1] [,2] [,3]
[1,] -2 2 -3
[2,] -1 1 3
[3,] 2 0 -1
> det(x)
[1] 18
> determinant(x)
$modulus
[1] 2.890372
attr(,"logarithm")
[1] TRUE
$sign
[1] 1
attr(,"class")
[1] "det"
dget Function
dput() and dget() function write or read an ASCII text representation of an R object to a file or connection, or uses one to recreate the object.
dput(x, file = "",
control = c("keepNA", "keepInteger", "showAttributes"))
dget(file)
x: R object
file: the file
control: character vector indicating deparsing options
...
diag Function
diag() function extracts or replaces the diagonal of a matrix, or constructs a diagonal matrix.
diag(x = 1, nrow, ncol)
diag(x) <- value
x: matrix, vector
nrow, ncol: Optional dimensions for the result when x is not a matrix
: either a single value or a vector of length equal to that of the current diagonal. Should be of a mode which can be coerced to that of x
...
> diag(10,3,4)
[,1] [,2] [,3] [,4]
[1,] 10 0 0 0
[2,] 0 10 0 0
[3,] 0 0 10 0
> diag(3)
[,1] [,2] [,3]
[1,] 1 0 0
[2,] 0 1 0
[3,] 0 0 1
diff Function
diff() function returns suitably lagged and iterated differences.
diff(x, ...)
diff(x, lag = 1, differences = 1, ...)
x: a numeric vector or matrix containing the values to be differenced
: an integer indicating which lag to use
: an integer indicating the order of the difference
...
> diff(1:10)
[1] 1 1 1 1 1 1 1 1 1
> diff(c(2,6,3,49,5))
[1] 4 -3 46 -44
difftime Function
difftime() function calculates time differences between two times.
difftime(time1, time2, tz,
units = c("auto", "secs", "mins", "hours",
"days", "weeks"))
time1, time2: date-time, date objects
tz: an optional timezone specification to be used for the conversion, mainly for "POSIXlt" objects
units: Units in which the results are desired, can be abbreviated
...
> x <- "2013-06-12 19:18:05"
> y <- "2013-06-13 19:18:05"
> difftime(x,y)
Time difference of -1 days
> x <- "2013-06-12 19:18:05"
> y <- "2013-06-13 12:18:23"
> difftime(x,y)
Time difference of -1 days
> y <- "2013-06-13 12:18:23"
> difftime(x,y)
Time difference of -17.005 hours
> difftime(x,y,tz="EST")
Time difference of -17.005 hours
digamma Function
digamma() function returns the first and second derivatives of the logarithm of the gamma function.
digamma(x) = Γ'(x)/Γ(x)
digamma(x)
x: numeric vector
> x <- c(2,6,3,49,5)
> digamma(x)
[1] 0.4227843 1.7061177 0.9227843 3.8815815 1.5061177
dim Function
dim() function gets or sets the dimension of a matrix, array or data frame.
dim(x)
x: array, matrix or data frame.
>BOD #R Biochemical Oxygen Demand Dataset
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
>class(BOD)
[1] "data.frame"
>dim(BOD) #get dimension
[1] 6 2
Set dimension of a matrix:
>x <- rep(1:20)
>x
[1] 1 2 3 4 5 6 7 8 9 10
Set dimension to 2 × 5:
>dim(x) <- c(2,5)
>x
[,1] [,2] [,3] [,4] [,5]
[1,] 1 3 5 7 9
[2,] 2 4 6 8 10
dimnames Function
dimnames() function retrieve or set the dimnames of an object.
dimnames(x)
dimnames(x) <- value
x: matrix, array or data frame
value: value for dimnames
...
> x <- read.csv("matrix.csv",header=T,sep="\t")
> dimnames(x)
[[1]]
[1] "1" "2" "3" "4" "5" "6" "7" "8" "9"
[[2]]
[1] "Subtype" "Expression" "Quality" "Height
dir Function
dir() function lists all the files in a directory.
dir(path = ".", pattern = NULL, all.files = FALSE,
full.names = FALSE, recursive = FALSE,
ignore.case = FALSE, include.dirs = FALSE)
path: a character vector of full path names; the default corresponds to the working directory, getwd(). Tilde expansion (see path.expand) is performed. Missing values will be ignored.
pattern: an optional regular expression. Only file names which match the regular expression will be returned
all.files: a logical value. If FALSE, only the names of visible files are returned. If TRUE, all file names will be returned
full.names: a logical value. If TRUE, the directory path is prepended to the file names to give a relative file path. If FALSE, the file names (rather than paths) are returned
recursive: logical. Should the listing recurse into directories?
ignore.case: logical. Should pattern-matching be case-insensitive?
include.dirs: logical. Should subdirectory names be included in recursive listings? (There always are in non-recursive ones)
dirname Function
dirname() function gets the directory name of a file.
dirname(x)
x: path name
> x <- "/usr/local/r/test.R"
> dirname(x)
[1] "/usr/local/r"
double Function
double() function creates a double-precision vector with default value 0.
double(length = 0)
as.double(x, ...)
is.double(x)
length: A non-negative integer specifying the desired length. Double values will be coerced to integer: supplying an argument of length other than one will give a warning
x: object to be coerced or tested
...
> x <- double(5)
> x
[1] 0 0 0 0 0
Quote Text Function
dQuote() function double quote text, sQuote()
function single quote text.
sQuote(x)
dQuote(x)
x: string, character vector
...
> x <- "2013-06-12 19:18:05"
> sQuote(x)
[1] "‘2013-06-12 19:18:05’"
> dQuote(x)
[1] "“2013-06-12 19:18:05”"
drop Function
drop() function delete the dimensions of an array which have only
one level.
drop(x)
x: array, matrix
droplevels Function
droplevels() function drop unused levels from factor.
droplevels(x,...)
droplevels(x, except, ...)
x: factor
except: indices of columns which not to drop levels
...
dump Function
dump() function takes a vector of names of R objects and produces text representations of the objects on a file or connection. A dump file can usually be sourced into another R (or S) session.
dump(list, file = "dumpdata.R", append = FALSE,
control = "all", envir = parent.frame(), evaluate = TRUE)
list: The names of one or more R objects to be dumped
file: either a character string naming a file or a connection. "" indicates output to the console
append: if TRUE and file is a character string, output will be appended to file; otherwise, it will overwrite the contents of file
control: character vector indicating deparsing options
envir: the environment to search for objects
evaluate: logical. Should promises be evaluated?
anyDuplicated Function
anyDuplicated() function determines which elements are duplicates in a vector or data frame.
duplicated(x, incomparables = FALSE, ...)
x: vector or data frame
incomparables: a vector of values that cannot be compared
fromlast: calculate from the vector end
...
> x <- c(1:5, 3:8, 7,8)
> x
[1] 1 2 3 4 5 3 4 5 6 7 8 7 8
> duplicated(x)
[1] FALSE FALSE FALSE FALSE FALSE TRUE TRUE TRUE FALSE FALSE
[11] FALSE TRUE TRUE
> x2 <- x[!duplicated(x)]
> x2
[1] 1 2 3 4 5 6 7 8
eapplay Function
eapplay() function applies function to the named values from an environment and returns the results as a list.
eapply(env, FUN, ..., all.names = FALSE, USE.NAMES = TRUE)
env: environment to be used
FUN: function to be applied
...: optional arguments to FUN
all.names: a logical indicating whether to apply the function to all values
USE.NAMES: logical indicating whether the resulting list should have names
...
> ev <- new.env(hash = FALSE)
> ev
<environment: 0x0000000010f1d7d0>
> ev$x <- c(4,9)
> eapply(ev,cumsum)
$x
[1] 4 13
eigen Function
eigen() function calculates eigenvalues and eigenvectors of matrices.
eigen(x, symmetric, only.values = FALSE, EISPACK = FALSE)
x: matrix
symmetric: if TRUE, the matrix is assumed to be symmetric (or Hermitian if complex) and only its lower triangle (diagonal included) is used. If symmetric is not specified, the matrix is inspected for symmetry
only.values: if TRUE, only the eigenvalues are computed and returned, otherwise both eigenvalues and eigenvectors are returned
EISPACK: logical. Should EISPACK be used (for compatibility with R < 1.7.0)?
> x <- matrix(1:9,3,3)
>x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> eigen(x)
$values
[1] 1.611684e+01 -1.116844e+00 -5.700691e-16
$vectors
[,1] [,2] [,3]
[1,] -0.4645473 -0.8829060 0.4082483
[2,] -0.5707955 -0.2395204 -0.8164966
[3,] -0.6770438 0.4038651 0.4082483
> x <- diag(6,4,4)
> x
[,1] [,2] [,3] [,4]
[1,] 6 0 0 0
[2,] 0 6 0 0
[3,] 0 0 6 0
[4,] 0 0 0 6
> eigen(x)
$values
[1] 6 6 6 6
$vectors
[,1] [,2] [,3] [,4]
[1,] 0 0 0 1
[2,] 0 0 1 0
[3,] 0 1 0 0
[4,] 1 0 0 0
encodeString Function
encodeString() function escapes the strings in a character vector in the same way print.default does, and optionally fits the encoded strings within a field width.
encodeString(x, width = 0, quote = "", na.encode = TRUE,
justify = c("left", "right", "centre", "none"))
x: string, character vector
width: integer: the minimum field width. If NULL or NA, this is taken to be the largest field width needed for any element of x
quote: character: quoting character, if any
na.encode: logical: should NA strings be encoded?
justify: character: partial matches are allowed. If padding to the minimum field width is needed, how should spaces be inserted? justify == "none" is equivalent to width = 0, for consistency with format.default
> x <- "r tutorial"
> encodeString(x)
[1] "r tutorial"
> x <- "r tutorial\n"
> encodeString(x)
[1] "r tutorial\\n"
Encoding Function
Encoding() function reads or sets the declared encodings for a character vector.
Encoding(x)
Encoding(x) <- value
enc2native(x)
enc2utf8(x)
x: string, character vector
value: string, character vector
> x <- "r tutorial"
> Encoding(x)
[1] "unknown"
> x <- "fa\xE7ile"
> Encoding(x)
[1] "latin1"
enquote Function
enquote() function is a simple one-line utility which transforms a call of the form Foo(....) into the call quote(Foo(....)). This is typically used to protect a call from early evaluation.
enquote(cl)
cl: a call
> enquote(lm)
quote(function (formula, data, subset, weights, na.action, method = "qr",
model = TRUE, x = FALSE, y = FALSE, qr = TRUE, singular.ok = TRUE,
contrasts = NULL, offset, ...)
{
ret.x <- x
ret.y <- y
cl <- match.call()
mf <- match.call(expand.dots = FALSE)
m <- match(c("formula", "data", "subset", "weights", "na.action",
"offset"), names(mf), 0L)
mf <- mf[c(1L, m)]
mf$drop.unused.levels <- TRUE
mf[[1L]] <- as.name("model.frame")
mf <- eval(mf, parent.frame())
if (method == "model.frame") return(mf) else if (method !=
"qr") warning(gettextf("method = '%s' is not supported.
Using 'qr'", method), domain = NA)
mt <- attr(mf, "terms")
y <- model.response(mf, "numeric")
w <- as.vector(model.weights(mf))
if (!is.null(w) && !is.numeric(w)) stop("'weights' must be
a numeric vector")
offset <- as.vector(model.offset(mf))
if (!is.null(offset)) {
if (length(offset) != NROW(y)) stop(gettextf("number of
offsets is %d, should equal %d (number of
observations)", length(offset), NROW(y)), domain = NA)
}
if (is.empty.model(mt)) {
x <- NULL
z <- list(coefficients = if (is.matrix(y)) matrix(, 0,
3) else numeric(), residuals = y, fitted.values = 0 *
y, weights = w, rank = 0L, df.residual = if (!is.null(w))
sum(w != 0) else if (is.matrix(y)) nrow(y) else length(y))
if (!is.null(offset)) {
z$fitted.values <- offset
z$residuals <- y - offset
}
} else {
x <- model.matrix(mt, mf, contrasts)
z <- if (is.null(w)) lm.fit(x, y, offset = offset,
singular.ok = singular.ok, ...)
else lm.wfit(x, y, w, offset = offset,
singular.ok = singular.ok, ...)
}
class(z) <- c(if (is.matrix(y)) "mlm", "lm")
z$na.action <- attr(mf, "na.action")
z$offset <- offset
z$contrasts <- attr(x, "contrasts")
z$xlevels <- .getXlevels(mt, mf)
z$call <- cl
z$terms <- mt
if (model) z$model <- mf
if (ret.x) z$x <- x
if (ret.y) z$y <- y
if (!qr) z$qr <- NULL
z
})
environment
environment(fun = NULL)
environment(fun) <- value
is.environment(x)
.GlobalEnv
globalenv()
.BaseNamespaceEnv
emptyenv()
baseenv()
new.env(hash = TRUE, parent = parent.frame(), size = 29L)
parent.env(env)
parent.env(env) <- value
environmentName(env)
env.profile(env)
fun: a function, a formula, or NULL, which is the default
value: an environment to associate with the function
x: an arbitrary R object
hash: a logical, if TRUE the environment will use a hash table
parent: an environment to be used as the enclosure of the environment created
env: an environment
size: an integer specifying the initial size for a hashed environment. An internal default value will be used if size is NA or zero. This argument is ignored if hash is FALSE
Binding and Environment Adjustments
These functions represent an experimental interface for adjustments to environments and bindings within environments. They allow for locking environments as well as individual bindings, and for linking a variable to a function.
lockEnvironment(env, bindings = FALSE)
environmentIsLocked(env)
lockBinding(sym, env)
unlockBinding(sym, env)
bindingIsLocked(sym, env)
makeActiveBinding(sym, fun, env)
bindingIsActive(sym, env)
env: an environment
bindings: logical specifying whether bindings should be locked
sym: a name object or character string
fun: a function taking zero or one arguments
eval Function
eval() function evaluates an R expression in a specified environment.
eval(expr, envir = parent.frame(),
enclos = if(is.list(envir) || is.pairlist(envir))
parent.frame() else baseenv())
evalq(expr, envir, enclos)
eval.parent(expr, n = 1)
local(expr, envir = new.env())
expr: an object to be evaluated
envir: the environment in which expr is to be evaluated. May also be NULL, a list, a data frame, a pairlist or an integer as specified to sys.call
enclos: Relevant when envir is a (pair)list or a data frame. Specifies the enclosure, i.e., where R looks for objects not found in envir. This can be NULL (interpreted as the base package environment, baseenv()) or an environment
n: number of parent generations to go back
> x <- 5
> eval(x * 3)
[1] 15
> eval(sin(pi/2))
[1] 1
exists Function
exists() function looks for an R object of the given name.
exists(x, where = -1, envir = , frame, mode = "any", inherits = TRUE)
x: variable
where: where to look for the object (see the details section); if omitted, the function will search as if the name of the object appeared unquoted in an expression
envir: an alternative way to specify an environment to look in, but it is usually simpler to just use the where argument
frame: a frame in the calling list. Equivalent to giving where as sys.frame(frame)
mode: the mode or type of object sought: see the ‘Details’ section
inherits: should the enclosing frames of the environment be searched?
> exists("lm")
[1] TRUE
> exists("sin")
[1] TRUE
exp Function
exp(x) function compute the exponential value of a number or number vector, ex.
> x <- 5
> exp(x)
[1] 148.4132
> y <- rep(1:20)
> exp(y)
[,1] [,2] [,3] [,4] [,5]
[1,] 2.718282 20.08554 148.4132 1096.633 8103.084
[2,] 7.389056 54.59815 403.4288 2980.958 22026.466
^ operator calculates a raised to power b:
> 2^3
[1] 8
> 4 ^ (1/2)
[1] 2
expm1() function computes exp() minus 1:
> expm1(5)
[1] 147.4132
> expm1(rep(1:20))
[,1] [,2] [,3] [,4] [,5]
[1,] 1.718282 19.08554 147.4132 1095.633 8102.084
[2,] 6.389056 53.59815 402.4288 2979.958 22025.466
Let's plot the exponential value in the range of -1 ~ 5:
> plot(exp(c(-1,0,0.2,0.3,1,2,3,4,5)),col="darkgreen")

expand.grid Function
expand.grid() function creates a data frame from all combinations of the supplied vectors or factors.
expand.grid(..., KEEP.OUT.ATTRS = TRUE, stringsAsFactors = TRUE)
...: vectors, factors or a list containing these
KEEP.OUT.ATTRS: a logical indicating the "out.attrs" attribute (see below) should be computed and returned
stringsAsFactors: logical specifying if character vectors are converted to factors
...
> subtype <- c("green","red","yellow")
> height <- c("3.2","2.5","6.1")
> sex <- c("M","F","F")
> expand.grid(subtype,height,sex)
Var1 Var2 Var3
1 green 3.2 M
2 red 3.2 M
3 yellow 3.2 M
4 green 2.5 M
5 red 2.5 M
6 yellow 2.5 M
7 green 6.1 M
8 red 6.1 M
9 yellow 6.1 M
10 green 3.2 F
11 red 3.2 F
12 yellow 3.2 F
13 green 2.5 F
14 red 2.5 F
15 yellow 2.5 F
16 green 6.1 F
17 red 6.1 F
18 yellow 6.1 F
19 green 3.2 F
20 red 3.2 F
21 yellow 3.2 F
22 green 2.5 F
23 red 2.5 F
24 yellow 2.5 F
25 green 6.1 F
26 red 6.1 F
27 yellow 6.1 F
> expand.grid(subtype,height)
Var1 Var2
1 green 3.2
2 red 3.2
3 yellow 3.2
4 green 2.5
5 red 2.5
6 yellow 2.5
7 green 6.1
8 red 6.1
9 yellow 6.1
expm1 Function
expm1(x) function calculates exp(x) - 1.
expm1(x)
x: Numeric or complex vector
> expm1(2)
[1] 6.389056
> expm1(1)
[1] 1.718282
> expm1(0)
[1] 0
> expm1(10)
[1] 22025.47
expression Function
expression() function creates or tests an R expression.
expression(...)
is.expression(x)
as.expression(x, ...)
...: R expression, like calls, symbols, constants
x: R object
> x <- expression(sin(pi/2))
> x
expression(sin(pi/2))
> eval(x)
[1] 1
> x <- "sin(pi/2)"
> x
[1] "sin(pi/2)"
> eval(x)
[1] "sin(pi/2)"
factor Function
R factors variable is a vector of categorical data. factor() function creates a factor variable, and calculates the categorical distribution of a vector data.
factor(x = character(), levels, labels = levels,
exclude = NA, ordered = is.ordered(x))
x: a vector of data
...
> v <- c(1,3,5,8,2,1,3,5,3,5)
> is.factor(v)
[1] FALSE
Calculates the categorical distribution:
> factor(v)
[1] 1 3 5 8 2 1 3 5 3 5
Levels: 1 2 3 5 8
> x <- factor(v)
> x
[1] 1 3 5 8 2 1 3 5 3 5
Levels: 1 2 3 5 8
> is.factor(x)
[1] TRUE
Select levels:
> x <- factor(v, levels=c(2,1))
> x
[1] 1 <NA> <NA> <NA> 2 1 <NA> <NA> <NA> <NA>
Levels: 2 1
Change the level value:
> levels(x) <- c("two","one")
> x
[1] one <NA> <NA> <NA> two one <NA> <NA> <NA> <NA>
Levels: two one
factorial Function
factorial() function computes the factorial of a number.
factorial(x)
x: numeric vector
> factorial(2) #2 × 1
[1] 2
> factorial(1) #1 × 1
[1] 1
> factorial(3) #3 × 2 × 1
[1] 6
> factorial(4) #4 × 3 × 2 × 1
[1] 24
> factorial(c(4,3,2))
[1] 24 6 2
bzfile Function
file() function open a file.
file(description = "", open = "", blocking = TRUE,
encoding = getOption("encoding"), raw = FALSE)
description: file name or connection.
open: open file mode.
encoding: the name of the encoding to be used.
...
> writ <- file("wfile.csv","w")
> cat("test ...",file=writ,sep="\n")
> close(writ)
find.package Function
find.package() function finds paths of packages.
find.package(package, lib.loc = NULL, quiet = FALSE,
verbose = getOption("verbose"))
path.package(package, quiet = FALSE)
library() #List all installed packages
package: name of package
lib.loc: a character vector describing the location of R library trees to search through, or NULL. The default value of NULL corresponds to checking the attached packages, then all libraries currently known in .libPaths()
quiet: logical. Should this not give warnings or an error if the package is not found?
verbose: logical. If TRUE, additional diagnostics are printed.
findInterval Function
findInterval(x,vec) function finds the indices of x in vec.
findInterval(x, vec, rightmost.closed = FALSE, all.inside = FALSE)
x: number
vec: numeric vector
rightmost.closed: logical; if true, the rightmost interval, vec[N-1] .. vec[N] is treated as closed
all.inside: logical; if true, the returned indices are coerced into 1,...,N-1, i.e., 0 is mapped to 1 and N to N-1
> v <- c(1:10)
> findInterval(3,v)
[1] 3
> v <- c(3,5,9,2,5,32,11)
> findInterval(9,v)
Error in findInterval(9, v) : 'vec' must be sorted non-decreasingly
> v2 <- sort(v)
> v2
[1] 2 3 5 5 9 11 32
> findInterval(9,v2)
[1] 5
> findInterval(5,v2)
[1] 4
finite Function
is.finite() and is.infinite() functions return a vector of the same length as x, indicating which elements are finite (not infinite and not missing) or infinite.
Inf and -Inf are positive and negative infinity whereas NaN means ‘Not a Number’. (These apply to numeric values and real and imaginary parts of complex values but not to values of integer vectors.) Inf and NaN are reserved words in the R language.
is.finite(x)
is.infinite(x)
is.nan(x)
x: the variable to be tested
> is.finite(3)
[1] TRUE
> x <- 3/0
> x
[1] Inf
> is.finite(x)
[1] FALSE
> is.infinite(x)
[1] TRUE
floor Function
floor() function returns the largest integer not greater than the giving number.
floor(x)
x: numeric
> floor(2.3)
[1] 2
> floor(2)
[1] 2
> floor(-3.2)
[1] -4
flush Function
flush() function flushes the output stream of a connection open for write/append.
force Function
force() function forces the evaluation of a function argument.
force(x)
x: a formal argument of the enclosing function
Foreign Function
R functions to make calls to compiled code that has been loaded into R,
including C, Fortran.
.C(.NAME, ..., NAOK = FALSE, DUP = TRUE, PACKAGE, ENCODING)
.Fortran(.NAME, ..., NAOK = FALSE, DUP = TRUE, PACKAGE, ENCODING)
.External(.NAME, ..., PACKAGE)
.Call(.NAME, ..., PACKAGE)
For Loop Example
Unlike other program languages, the for loop of R language can be write as for (i in arr) {expr1; expr2 ...}.
It goes through the vector arr every time one element i, and execute a group of
commands inside the { ... } in each cycle. The break statement can be used to terminate the loop abruptly.
If you don't want to terminate the whole loop, but just ignore current cycle, the next statement can do that.
Let's create a vector containing number 1-10:
>samples <- c(rep(1:10))
>samples
[1] 1 2 3 4 5 6 7 8 9 10
Go through the samples one by one and print them out:
>for (thissample in samples)
+{
+ print(thissample)
+}
[1] 1
[1] 2
[1] 3
[1] 4
[1] 5
[1] 6
[1] 7
[1] 8
[1] 9
[1] 10
Let's do something inside the for loop:
>for (thissample in samples)
+{
+ str <- paste(thissample,"is current sample",sep=" ")
+ print(str)
+}
[1] "1 is current sample"
[1] "2 is current sample"
[1] "3 is current sample"
[1] "4 is current sample"
[1] "5 is current sample"
[1] "6 is current sample"
[1] "7 is current sample"
[1] "8 is current sample"
[1] "9 is current sample"
[1] "10 is current sample"
Let's terminate the loop when the sample is 3:
>for (thissample in samples)
+{
+ if (thissample == 3) break
+ str <- paste(thissample,"is current sample",sep=" ")
+ print(str)
+}
[1] "1 is current sample"
[1] "2 is current sample"
Let's ignore when the sample number is even:
>for (thissample in samples)
+{
+ if (thissample %% 2 == 0) next
+ str <- paste(thissample,"is current sample",sep=" ")
+ print(str)
+}
[1] "1 is current sample"
[1] "3 is current sample"
[1] "5 is current sample"
[1] "7 is current sample"
[1] "9 is current sample"
Let's just loop through last three samples:
>end <- length(samples)
>begin <- end - 2
>for (thissample in begin:end)
+{
+ str <- paste(thissample,"is current sample",sep=" ")
+ print(str)
+}
[1] "8 is current sample"
[1] "9 is current sample"
[1] "10 is current sample"
formals Function
formals() function gets or sets the formal arguments of a function.
formals(fun = sys.function(sys.parent()))
formals(fun, envir = environment(fun)) <- value
fun: the function
envir: environment of the function
value: list of R expressions
> formals(dim)
NULL
> formals(split)
$x
$f
$drop
[1] FALSE
$...
format Function
format() function formats an R object for pretty printing.
format(x, trim = FALSE, digits = NULL, nsmall = 0L,
justify = c("left", "right", "centre", "none"),
width = NULL, na.encode = TRUE, scientific = NA,
big.mark = "", big.interval = 3L,
small.mark = "", small.interval = 5L,
decimal.mark = ".", zero.print = NULL,
drop0trailing = FALSE, ...)
x: any R object (conceptually); typically numeric.
trim: logical; if FALSE, logical, numeric and complex values are right-justified to a common width: if TRUE the leading blanks for justification are suppressed.
digits: how many significant digits are to be used for numeric and complex x. The default, NULL, uses getOption(digits). This is a suggestion: enough decimal places will be used so that the smallest (in magnitude) number has this many significant digits, and also to satisfy nsmall. (For the interpretation for complex numbers see signif.)
nsmall: the minimum number of digits to the right of the decimal point in formatting real/complex numbers in non-scientific formats. Allowed values are 0 <= nsmall <= 20.
justify: should a character vector be left-justified (the default), right-justified, centred or left alone.
width: default method: the minimum field width or NULL or 0 for no restriction.
AsIs method: the maximum field width for non-character objects. NULL corresponds to the default 12.
na.encode: logical: should NA strings be encoded? Note this only applies to elements of character vectors, not to numerical or logical NAs, which are always encoded as "NA".
scientific: Either a logical specifying whether elements of a real or complex vector should be encoded in scientific format, or an integer penalty (see options("scipen")). Missing values correspond to the current default penalty.
...: further arguments passed to or from other methods.
big.mark, big.interval, small.mark, small.interval, decimal.mark, zero.print, drop0trailing: used for prettying (longish) decimal sequences, passed to prettyNum: that help page explains the details.
> format(pi,digits=4)
[1] "3.142"
> format(pi,digits=4,nsmall=5)
[1] "3.14159"
forwardsolve Function
forwardsolve() function solves a system of linear
equations where the coefficient matrix is lower triangular.
x <- forwardsolve(L, b)
forwardsolve(l, x, k=ncol(l), upper.tri=FALSE, transpose=FALSE)
l: lower triangular matrix
x: a matrix whose columns give the right-hand sides for the equations
k: The number of columns of r and rows of x to use
F-test Example
var.test() function performs F-test between 2 normal populations with hypothesis that variances of the 2 populations are equal.
var.test(x, ...)
var.test(x, y, ratio = 1, alternative = c("two.sided", "less", "greater"),
conf.level = 0.95, ...)
x,y: Normally distributed data sets
ratio: Hypothesized ratio of x/y, default is 1
alternative: alternative hypothesis, including "two.sided","greater","less"
conf.level: confidence level
...
> x <- rnorm(100, mean=0)
> y <- rnorm(100, mean=1)
> var.text(x,y)
F test to compare two variances
data: x and y
F = 0.8795, num df = 99, denom df = 99, p-value = 0.5242
alternative hypothesis: true ratio of variances is not equal to 1
95 percent confidence interval:
0.5917706 1.3071567
sample estimates:
ratio of variances
0.8795095
Since the p-value = 0.5242, which is much higher than 0.05, the hypothesis that the variances of x and y are equal
is accepted.
gamma Function
gamma() function returns the gamma function Γx.
gamma(x): numeric vectors
> gamma(2)
[1] 1
> gamma(3)
[1] 2
> gamma(4)
[1] 6
> gamma(5)
[1] 24
> gamma(6)
[1] 120
> gamma(c(4,5,6))
[1] 6 24 120
gc Function
gc() function starts a garbage collection. gcinfo() sets a flag so that automatic collection is either silent (verbose=FALSE) or prints memory usage statistics (verbose=TRUE).
gc(verbose = getOption("verbose"), reset=FALSE)
gcinfo(verbose)
verbose: logical; if TRUE, the garbage collection prints statistics about cons cells and the space allocated for vectors
reset: logical; if TRUE the values for maximum space used are reset to the current values
> gc() #start garbage collection
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 204746 11.0 407500 21.8 350000 18.7
Vcells 313545 2.4 786432 6.0 786079 6.0
> gcinfo(TRUE)
[1] FALSE
> gc(TRUE)
Garbage collection 95 = 83+8+4 (level 2) ...
11.0 Mbytes of cons cells used (50%)
2.4 Mbytes of vectors used (40%)
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 204761 11.0 407500 21.8 350000 18.7
Vcells 313574 2.4 786432 6.0 786079 6.0
> gc.time()
[1] 0 0 0 0 0
Garbage Collection
gctorture() function provokes garbage collection on (nearly) every memory allocation. Intended to ferret out memory protection bugs. Also makes R run very slowly.
gctorture(on = TRUE)
gctorture2(step, wait = step, inhibit_release = FALSE)
on: turning on or off
step: run GC every step allocations; step = 0 turns the GC torture off
wait: number of allocations to wait before starting GC torture
inhibit_release: do not release free objects for re-use: use with caution
get Function
get() function searches for an R object with a given name and return it.
get(x, pos = -1, envir = as.environment(pos), mode = "any",
inherits = TRUE)
mget(x, envir, mode = "any",
ifnotfound = list(function(x)
stop(paste("value for '", x, "' not found", sep = ""),
call. = FALSE)),
inherits = FALSE)
x: the variable
pos: where to look for the object (see the details section); if omitted, the function will search as if the name of the object appeared unquoted in an expression
envir: an alternative way to specify an environment to look in; see the ‘Details’ section
mode: the mode or type of object sought: see the ‘Details’ section
inherits: should the enclosing frames of the environment be searched?
ifnotfound: A list of values to be used if the item is not found: it will be coerced to list if necessary
> x <- 5
> get("x")
[1] 5
geterrmessage Function
geterrmessage() function gets the last error message.
getLoadedDLLs Function
getLoadedDLLs() function gets a list of all the DLLs that are currently loaded.
getNativeSymbolInfo Function
getNativeSymbolInfo() function finds and returns as comprehensive a description of one or more dynamically loaded or ‘exported’ built-in native symbols. For each name, it returns information about the name of the symbol, the library in which it is located and, if available, the number of arguments it expects and by which interface it should be called (i.e .Call, .C, .Fortran, or .External). Additionally, it returns the address of the symbol and this can be passed to other C routines which can invoke. Specifically, this provides a way to explicitly share symbols between different dynamically loaded package libraries. Also, it provides a way to query where symbols were resolved, and aids diagnosing strange behavior associated with dynamic resolution.
getNativeSymbolInfo(name, PACKAGE, unlist = TRUE,
withRegistrationInfo = FALSE)
name: the name(s) of the native symbol(s) as used in a call to is.loaded, etc. Note that Fortran symbols should be supplied as-is, not wrapped in symbol.For.
PACKAGE: an optional argument that specifies to which DLL we restrict the search for this symbol. If this is "base", we search in the R executable itself
unlist: a logical value which controls how the result is returned if the function is called with the name of a single symbol. If unlist is TRUE and the number of symbol names in name is one, then the NativeSymbolInfo object is returned. If it is FALSE, then a list of NativeSymbolInfo objects is returned. This is ignored if the number of symbols passed in name is more than one. To be compatible with earlier versions of this function, this defaults to TRUE
withRegistrationInfo: a logical value indicating whether, if TRUE, to return information that was registered with R about the symbol and its parameter types if such information is available, or if FALSE to return the address of the symbol
getOption Function
getOption() function allows the user to set and examine a variety of global options which affect the way in which R computes and displays its results.
getOption(x, default = NULL)
x: character string holding an option name.
...
References to Source Files
srcfile(filename, encoding = getOption("encoding"), Enc = "unknown")
srcfilecopy(filename, lines)
getSrcLines(srcfile, first, last)
srcref(srcfile, lloc)
# S3 method for class 'srcfile'
print(x, ...)
# S3 method for class 'srcfile'
summary(object, ...)
# S3 method for class 'srcfile'
open(con, line, ...)
# S3 method for class 'srcfile'
close(con, ...)
# S3 method for class 'srcref'
print(x, useSource = TRUE, ...)
# S3 method for class 'srcref'
summary(object, useSource = FALSE, ...)
# S3 method for class 'srcref'
as.character(x, useSource = TRUE, ...)
.isOpen(srcfile)
filename: the file name
encoding: encoding of the file
Enc: the encoding with which to make strings
lines: A character vector of source lines. Other R objects will be coerced to character
srcfile: srcfile object
first, last, line: line numbers
lloc: vector of four, six or eight values giving a source location; see ‘Details’
x, object, con: an object of the appropriate class
useSource: whether to read the srcfile to obtain the text of a srcref
...
Let's see a source file "tp.R":
> src <- srcfile("tp.R")
> getSrcLines(src,1,3)
> lines <- getSrcLines(src,1,3)
> lines
[1] "function sum(a,b)" "{" " x <- a + b"
gettext Function
gettext(..., domain = NULL)
ngettext(n, msg1, msg2, domain = NULL)
bindtextdomain(domain, dirname = NULL)
...: character vectors
domain: domain of translation
n: non-negative integer
msg1: the message to be used in English for n = 1
msg2: the message to be used in English for n = 0, 2, 3,...
dirname: the directory in which to find translated message catalogs for the domain
getwd Function
getwd() function returns the current R working directory.
setwd() function sets the current R working directory.
getwd()
setwd(dir)
dir: the directory to be set
...
> getwd()
[1] "/user/r"
> setwd("/user/")
> getwd("")
[1] "/user/"
> setwd("../")
> getwd()
[1] "/"
gl Function
gl() function generates factors by specifying the pattern of their levels.
gl(n, k, length = n*k, labels = 1:n, ordered = FALSE)
n: number of levels
k: number of replications
length: length of the result
labels: labels for the resulting factor levels
ordered: whether the result sould be ordered or not
> gl(3,2,labels = c("green","red","yellow"))
[1] green green red red yellow yellow
Levels: green red yellow
glm Function
glm() function fits linear models to the dataset.
glm(formula, family = gaussian, data, weights, subset,
na.action, start = NULL, etastart, mustart, offset,
control = list(...), model = TRUE, method = "glm.fit",
x = FALSE, y = TRUE, contrasts = NULL, ...)
>Orange #R growth of orange trees dataset
Tree age circumference
1 1 118 30
2 1 484 58
3 1 664 87
4 1 1004 115
5 1 1231 120
6 1 1372 142
7 1 1582 145
8 2 118 33
9 2 484 69
10 2 664 111
11 2 1004 156
12 2 1231 172
13 2 1372 203
14 2 1582 203
15 3 118 30
16 3 484 51
17 3 664 75
18 3 1004 108
19 3 1231 115
20 3 1372 139
21 3 1582 140
22 4 118 32
23 4 484 62
24 4 664 112
25 4 1004 167
26 4 1231 179
27 4 1372 209
28 4 1582 214
29 5 118 30
30 5 484 49
31 5 664 81
32 5 1004 125
33 5 1231 142
34 5 1372 174
35 5 1582 177
> attach(Orange) #put age, Tree, circumference into R search path
> g <- glm(circumference ~ age + Tree)
> g
Call: glm(formula = circumference ~ age + Tree)
Coefficients:
(Intercept) age Tree.L Tree.Q Tree.C Tree^4
17.3997 0.1068 39.9350 2.5199 -8.2671 -4.6955
Degrees of Freedom: 34 Total (i.e. Null); 29 Residual
Null Deviance: 112400
Residual Deviance: 6754 AIC: 297.5
>summary(g)
Call:
glm(formula = circumference ~ age + Tree)
Deviance Residuals:
Min 1Q Median 3Q Max
-30.505 -8.790 3.737 7.650 21.859
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 17.399650 5.543461 3.139 0.00388 **
age 0.106770 0.005321 20.066 < 2e-16 ***
Tree.L 39.935049 5.768048 6.923 1.31e-07 ***
Tree.Q 2.519892 5.768048 0.437 0.66544
Tree.C -8.267097 5.768048 -1.433 0.16248
Tree^4 -4.695541 5.768048 -0.814 0.42224
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for gaussian family taken to be 232.8927)
Null deviance: 112366.3 on 34 degrees of freedom
Residual deviance: 6753.9 on 29 degrees of freedom
AIC: 297.51
Number of Fisher Scoring iterations: 2
gregexpr Function
• gregexpr returns a list of the same length as text each
element of which is of the same form as the return value for regexpr,
except that the starting positions of every (disjoint) match are
given.
gregexpr(pattern, text, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
•
pattern: regular expression, or string for fixed=TRUE
•
text: string, the character vector
•
ignore.case: case sensitive or not
•
perl: logical. Should perl-compatible regexps be used? Has priority over extended
•
fixed: logical. If TRUE, pattern is a string to be matched as is. Overrides all conflicting arguments
•
useBytes: logical. If TRUE the matching is done byte-by-byte rather than character-by-character
> x <- "line 4322: He is now 25 years old, and weights 130lbs"
> y <- gregexpr("\\d+",x)
> y
[[1]]
[1] 6 22 48
attr(,"match.length")
[1] 4 2 3
attr(,"useBytes")
[1] TRUE
> if (y[[1]][1] != -1) print("match")
[1] "match"
>str <- c("Regular", "expression", "examples of R language")
>x <- gregexpr("x*ress",str)
>x
[[1]]
[1] -1
attr(,"match.length")
[1] -1
attr(,"useBytes")
[1] TRUE
[[2]]
[1] 4
attr(,"match.length")
[1] 4
attr(,"useBytes")
[1] TRUE
[[3]]
[1] -1
attr(,"match.length")
[1] -1
attr(,"useBytes")
[1] TRUE
Regular Expression Syntax:
| Syntax | Description |
| \\d | Digit, 0,1,2 ... 9 |
| \\D | Not Digit |
| \\s | Space |
| \\S | Not Space |
| \\w | Word |
| \\W | Not Word |
| \\t | Tab |
| \\n | New line |
| ^ | Beginning of the string |
| $ | End of the string |
| \ | Escape special characters, e.g. \\ is "\", \+ is "+" |
| | | Alternation match. e.g. /(e|d)n/ matches "en" and "dn" |
| • | Any character, except \n or line terminator |
| [ab] | a or b |
| [^ab] | Any character except a and b |
| [0-9] | All Digit |
| [A-Z] | All uppercase A to Z letters |
| [a-z] | All lowercase a to z letters |
| [A-z] | All Uppercase and lowercase a to z letters |
| i+ | i at least one time |
| i* | i zero or more times |
| i? | i zero or 1 time |
| i{n} | i occurs n times in sequence |
| i{n1,n2} | i occurs n1 - n2 times in sequence |
| i{n1,n2}? | non greedy match, see above example |
| i{n,} | i occures >= n times |
| [:alnum:] | Alphanumeric characters: [:alpha:] and [:digit:] |
| [:alpha:] | Alphabetic characters: [:lower:] and [:upper:] |
| [:blank:] | Blank characters: e.g. space, tab |
| [:cntrl:] | Control characters |
| [:digit:] | Digits: 0 1 2 3 4 5 6 7 8 9 |
| [:graph:] | Graphical characters: [:alnum:] and [:punct:] |
| [:lower:] | Lower-case letters in the current locale |
| [:print:] | Printable characters: [:alnum:], [:punct:] and space |
| [:punct:] | Punctuation character: ! " # $ % & ' ( ) * + , - . / : ; < = > ? @ [ \ ] ^ _ ` { | } ~ |
| [:space:] | Space characters: tab, newline, vertical tab, form feed, carriage return, space |
| [:upper:] | Upper-case letters in the current locale |
| [:xdigit:] | Hexadecimal digits: 0 1 2 3 4 5 6 7 8 9 A B C D E F a b c d e f |
Regular Expression
R has various functions for regular expression based match and replaces. The grep, grepl, regexpr and gregexpr functions are used for searching
for matches, while sub and gsub for performing replacement.
• grep(value = FALSE) returns an integer vector of the indices
of the elements of x that yielded a match (or not, for
invert = TRUE).
>str <- c("Regular", "expression", "examples of R language")
>x <- grep("ex",str,value=F)
>x
[1] 2 3
>x <- "line 4322: He is now 25 years old, and weights 130lbs";
>x <- grep("\\d","",x)
>x
[1] 1
• grep(value = TRUE) returns a character vector containing the
selected elements of x (after coercion, preserving names but no
other attributes).
>x <- grep("ex",str,value=T)
>x
[1] "expression" "examples of R language"
• grepl returns a logical vector (match or not for each element of
x).
>x <- grepl("ex",str)
>x
[1] FALSE TRUE TRUE
• sub and gsub return a character vector of the same
length and with the same attributes as x (after possible
coercion to character). Elements of character vectors x which
are not substituted will be returned unchanged (including any declared
encoding). If useBytes = FALSE a non-ASCII substituted result
will often be in UTF-8 with a marked encoding (e.g. if there is a
UTF-8 input, and in a multibyte locale unless fixed = TRUE).
>str <- c("Regular", "expression", "examples of R language")
>x <- sub("x.ress","",str)
>x
[1] "Regular" "eion" "examples of R language"
>x <- sub("x.+e","",str)
>x
[1] "Regular" "ession" "e"
>x <- "line 4322: He is now 25 years old, and weights 130lbs";
>x <- gsub("[[:digit:]]","",x)
>x
[1] "line : He is now years old, and weights lbs"
>x <- "line 4322: He is now 25 years old, and weights 130lbs";
>x <- gsub("\\d+","",x)
>x
[1] "line : He is now years old, and weights lbs"
• regexpr returns an integer vector of the same length as
text giving the starting position of the first match or
-1 if there is none, with attribute "match.length", an
integer vector giving the length of the matched text (or -1 for
no match). The match positions and lengths are in characters unless
useBytes = TRUE is used, when they are in bytes.
>str <- c("Regular", "expression", "examples of R language")
>x <- regexpr("x*ress",str)
>x
[1] -1 4 -1
• gregexpr returns a list of the same length as text each
element of which is of the same form as the return value for regexpr,
except that the starting positions of every (disjoint) match are
given.
>str <- c("Regular", "expression", "examples of R language")
>x <- gregexpr("x*ress",str)
>x
[[1]]
[1] -1
attr(,"match.length")
[1] -1
attr(,"useBytes")
[1] TRUE
[[2]]
[1] 4
attr(,"match.length")
[1] 4
attr(,"useBytes")
[1] TRUE
[[3]]
[1] -1
attr(,"match.length")
[1] -1
attr(,"useBytes")
[1] TRUE
Function Syntax:
grep(pattern, x, ignore.case = FALSE, perl = FALSE, value = FALSE,
fixed = FALSE, useBytes = FALSE, invert = FALSE)
grepl(pattern, x, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
sub(pattern, replacement, x, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
gsub(pattern, replacement, x, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
regexpr(pattern, text, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
gregexpr(pattern, text, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
Regular Expression Syntax:
| Syntax | Description |
| \\d | Digit, 0,1,2 ... 9 |
| \\D | Not Digit |
| \\s | Space |
| \\S | Not Space |
| \\w | Word |
| \\W | Not Word |
| \\t | Tab |
| \\n | New line |
| ^ | Beginning of the string |
| $ | End of the string |
| \ | Escape special characters, e.g. \\ is "\", \+ is "+" |
| | | Alternation match. e.g. /(e|d)n/ matches "en" and "dn" |
| • | Any character, except \n or line terminator |
| [ab] | a or b |
| [^ab] | Any character except a and b |
| [0-9] | All Digit |
| [A-Z] | All uppercase A to Z letters |
| [a-z] | All lowercase a to z letters |
| [A-z] | All Uppercase and lowercase a to z letters |
| i+ | i at least one time |
| i* | i zero or more times |
| i? | i zero or 1 time |
| i{n} | i occurs n times in sequence |
| i{n1,n2} | i occurs n1 - n2 times in sequence |
| i{n1,n2}? | non greedy match, see above example |
| i{n,} | i occures >= n times |
| [:alnum:] | Alphanumeric characters: [:alpha:] and [:digit:] |
| [:alpha:] | Alphabetic characters: [:lower:] and [:upper:] |
| [:blank:] | Blank characters: e.g. space, tab |
| [:cntrl:] | Control characters |
| [:digit:] | Digits: 0 1 2 3 4 5 6 7 8 9 |
| [:graph:] | Graphical characters: [:alnum:] and [:punct:] |
| [:lower:] | Lower-case letters in the current locale |
| [:print:] | Printable characters: [:alnum:], [:punct:] and space |
| [:punct:] | Punctuation character: ! " # $ % & ' ( ) * + , - . / : ; < = > ? @ [ \ ] ^ _ ` { | } ~ |
| [:space:] | Space characters: tab, newline, vertical tab, form feed, carriage return, space |
| [:upper:] | Upper-case letters in the current locale |
| [:xdigit:] | Hexadecimal digits: 0 1 2 3 4 5 6 7 8 9 A B C D E F a b c d e f |
grepl Function
grepl returns TRUE if a string contains the pattern, otherwise FALSE; if the parameter is a string vector, returns a logical vector (match or not for each element of
the vector).
grepl(pattern, x, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
•
pattern: regular expression, or string for fixed=TRUE
•
x: string, the character vector
•
ignore.case: case sensitive or not
•
perl: logical. Should perl-compatible regexps be used? Has priority over extended
•
fixed: logical. If TRUE, pattern is a string to be matched as is. Overrides all conflicting arguments
•
useBytes: logical. If TRUE the matching is done byte-by-byte rather than character-by-character
> x <- "line 4322: He is now 25 years old, and weights 130lbs"
> y <- grepl("\\d+",x)
> y
[1] TRUE
> x <- "line 4322: He is now 25 years old, and weights 130lbs"
> y <- grepl("[[:digit:]]",x)
> y
[1] TRUE
Vector match:
>str <- c("Regular", "expression", "examples of R language")
>x <- grepl("x*ress",str)
>x
[1] FALSE TRUE FALSE
Regular Expression Syntax:
| Syntax | Description |
| \\d | Digit, 0,1,2 ... 9 |
| \\D | Not Digit |
| \\s | Space |
| \\S | Not Space |
| \\w | Word |
| \\W | Not Word |
| \\t | Tab |
| \\n | New line |
| ^ | Beginning of the string |
| $ | End of the string |
| \ | Escape special characters, e.g. \\ is "\", \+ is "+" |
| | | Alternation match. e.g. /(e|d)n/ matches "en" and "dn" |
| • | Any character, except \n or line terminator |
| [ab] | a or b |
| [^ab] | Any character except a and b |
| [0-9] | All Digit |
| [A-Z] | All uppercase A to Z letters |
| [a-z] | All lowercase a to z letters |
| [A-z] | All Uppercase and lowercase a to z letters |
| i+ | i at least one time |
| i* | i zero or more times |
| i? | i zero or 1 time |
| i{n} | i occurs n times in sequence |
| i{n1,n2} | i occurs n1 - n2 times in sequence |
| i{n1,n2}? | non greedy match, see above example |
| i{n,} | i occures >= n times |
| [:alnum:] | Alphanumeric characters: [:alpha:] and [:digit:] |
| [:alpha:] | Alphabetic characters: [:lower:] and [:upper:] |
| [:blank:] | Blank characters: e.g. space, tab |
| [:cntrl:] | Control characters |
| [:digit:] | Digits: 0 1 2 3 4 5 6 7 8 9 |
| [:graph:] | Graphical characters: [:alnum:] and [:punct:] |
| [:lower:] | Lower-case letters in the current locale |
| [:print:] | Printable characters: [:alnum:], [:punct:] and space |
| [:punct:] | Punctuation character: ! " # $ % & ' ( ) * + , - . / : ; < = > ? @ [ \ ] ^ _ ` { | } ~ |
| [:space:] | Space characters: tab, newline, vertical tab, form feed, carriage return, space |
| [:upper:] | Upper-case letters in the current locale |
| [:xdigit:] | Hexadecimal digits: 0 1 2 3 4 5 6 7 8 9 A B C D E F a b c d e f |
gsub Function
gsub() function replaces all matches of a string, if the parameter is a string vector, returns a string vector of the same length and with the same attributes
(after possible coercion to character). Elements of string vectors which are not substituted
will be returned unchanged (including any declared encoding).
gsub(pattern, replacement, x, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
•
pattern: string to be matched
•
replacement: string for replacement
•
x: string or string vector
•
ignore.case: if TRUE, ignore case
...
> x <- "R Tutorial"
> gsub("ut","ot",x)
[1] "R Totorial"
Case insensitive replace:
> gsub("tut","ot",x,ignore.case=T))
[1] "R otorial"
If ignore.case is not set to True, no replace take place:
> gsub("tut","ot",x)
[1] "R Tutorial"
> x <- "line 4322: He is now 25 years old, and weights 130lbs"
> y <- gsub("\\d+","---",x)
> y
[1] "line ---: He is now --- years old, and weights ---lbs"
> x <- "line 4322: He is now 25 years old, and weights 130lbs"
> y <- gsub("[[:lower:]]","-",x)
> y
[1] "---- 4322: H- -- --- 25 ----- ---, --- ------- 130---"
Vector replacement:
> x <- c("R Tutorial","PHP Tutorial", "HTML Tutorial")
> gsub("Tutorial","Examples",x)
[1] "R Examples" "PHP Examples" "HTML Examples"
Regular Expression Syntax:
| Syntax | Description |
| \\d | Digit, 0,1,2 ... 9 |
| \\D | Not Digit |
| \\s | Space |
| \\S | Not Space |
| \\w | Word |
| \\W | Not Word |
| \\t | Tab |
| \\n | New line |
| ^ | Beginning of the string |
| $ | End of the string |
| \ | Escape special characters, e.g. \\ is "\", \+ is "+" |
| | | Alternation match. e.g. /(e|d)n/ matches "en" and "dn" |
| • | Any character, except \n or line terminator |
| [ab] | a or b |
| [^ab] | Any character except a and b |
| [0-9] | All Digit |
| [A-Z] | All uppercase A to Z letters |
| [a-z] | All lowercase a to z letters |
| [A-z] | All Uppercase and lowercase a to z letters |
| i+ | i at least one time |
| i* | i zero or more times |
| i? | i zero or 1 time |
| i{n} | i occurs n times in sequence |
| i{n1,n2} | i occurs n1 - n2 times in sequence |
| i{n1,n2}? | non greedy match, see above example |
| i{n,} | i occures >= n times |
| [:alnum:] | Alphanumeric characters: [:alpha:] and [:digit:] |
| [:alpha:] | Alphabetic characters: [:lower:] and [:upper:] |
| [:blank:] | Blank characters: e.g. space, tab |
| [:cntrl:] | Control characters |
| [:digit:] | Digits: 0 1 2 3 4 5 6 7 8 9 |
| [:graph:] | Graphical characters: [:alnum:] and [:punct:] |
| [:lower:] | Lower-case letters in the current locale |
| [:print:] | Printable characters: [:alnum:], [:punct:] and space |
| [:punct:] | Punctuation character: ! " # $ % & ' ( ) * + , - . / : ; < = > ? @ [ \ ] ^ _ ` { | } ~ |
| [:space:] | Space characters: tab, newline, vertical tab, form feed, carriage return, space |
| [:upper:] | Upper-case letters in the current locale |
| [:xdigit:] | Hexadecimal digits: 0 1 2 3 4 5 6 7 8 9 A B C D E F a b c d e f |
gzcon Function
gzcon() function provides a modified connection that wraps an existing connection, and decompresses reads or compresses writes through that connection. Standard gzip headers are assumed.
gzcon(con, level = 6, allowNonCompressed = TRUE)
con: connection
level: integer between 0 and 9, the compression level when writing
allowNonCompressed: logical. When reading, should non-compressed input be allowed?
Heatmap Plot
Heatmap needs "ctc" package from bioconductor, to install "ctc" package:
>source("http://bioconductor.org/biocLite.R")
>biocLite("ctc")
heatmap(...) function can draw a heatmap, it's usage is:
heatmap(x, Rowv=NULL, Colv=if(symm)"Rowv" else NULL,
distfun = dist, hclustfun = hclust,
reorderfun = function(d,w) reorder(d,w),
add.expr, symm = FALSE, revC = identical(Colv, "Rowv"),
scale=c("row", "column", "none"), na.rm = TRUE,
margins = c(5, 5), ColSideColors, RowSideColors,
cexRow = 0.2 + 1/log10(nr), cexCol = 0.2 + 1/log10(nc),
labRow = NULL, labCol = NULL, main = NULL,
xlab = NULL, ylab = NULL,
keep.dendro = FALSE, verbose = getOption("verbose"), ...)
x: Numeric matrix
Rowv: Row dendrogram
Colv: Column dendrogram
...
Let's first have a look of our data file named heatmap.csv:
elements S1 S2 S3 S4 S5 S6 S7 S8
R1 -0.0027 0.1057 0.1976 0.0209 0 0.0089 0.0082 0.0209
R2 0 -0.1204 0.2627 0 0 0.283 0.2076 -0.0158
R3 0 -0.1204 0.2627 0 0 0.283 0.2076 -0.0158
R4 0.0142 0 -0.454 0.0101 -0.0213 -0.0084 -0.0121 0.0083
R5 0 0 -0.2334 0.007 0.4151 0 0.0987 0.021
R6 0.0381 0.0644 0.2302 0 0 -0.0476 0.2432 -0.0069
R7 0.0381 0.0644 0.2302 0 0 -0.0476 0.2432 -0.0069
R8 0.0381 0.0644 0.2302 0 0 -0.0476 0.2432 -0.0069
R9 0.0891 -0.1022 -0.4466 -0.4877 -0.0175 -0.0523 -0.4792 -0.0547
R10 0.0046 -0.1539 -0.4645 0 -0.0282 0 -0.0217 0.017
R11 0.0706 0.028 0.3626 0 0.0196 -0.0094 0.3086 0
R12 0.0311 0.0759 0.2119 0 -0.0022 0 0 0.0117
R13 0.0013 0.0702 -0.3176 0.0152 0.0095 -0.0224 0.2069 0.005
R14 0.0491 0.0525 -0.4329 0.0237 -0.0038 -0.0224 0.2065 0.005
R15 0.0256 0.0579 0.1846 0.0024 0.0029 -0.0165 0.4781 -0.0123
R16 -0.0061 -0.1554 -0.0635 0.0121 -0.0282 0 -0.016 0.017
R17 -0.0061 -0.1554 -0.0635 0.0121 -0.0282 0 -0.016 0.017
Let's draw a simple heatmap:
>x <- read.csv("heatmap.csv", header=T, dec=".",sep=",")
>imageVals <- as.matrix(cn[2:nrow(x),2:ncol(x)]);
>heatmap(imageVals)

For further improvement, we's like to replace the rownames at the right side with names in the first column of the file.
Suppose S1-S4 come from location A, and s5-S8 come from location B, we will marked it as red and blue color as a bar under
the top dendrogram.
>rowNames = x[,1];
>samplecolors <- c("red","red","red","red","blue","blue","blue","blue");
>heatmap(imageVals,labRow=rowNames,ColSideColors=as.vector(samplecolors))

hexmode Function
hexmode() function converts or prints integers in hexadecimal format, with as many digits as are needed to display the largest, using leading zeroes as necessary.
as.hexmode(x)
as.character(x, ...)
format(x, width = NULL, upper.case = FALSE, ...)
x: R object to be converted
...
> x <- 3
> as.hexmode(x)
[1] "3"
> x <- 145
> as.hexmode(x)
[1] "91"
Histogram Plot Example
Histogram is a popular descriptive statistical method that shows data by dividing the range of values into intervals and plotting the frequency/density per interval as a bar.
hist(x, breaks = "Sturges", freq = NULL, ...)
x: value vector
breaks: number of bars
...
Following is a csv file example "histogram.csv", we will draw a Histogram of "Expression" values:
Let first read in the data from the file:
>x <- read.csv("histogram.csv",header=T,sep="\t")
>x <- t(x)
>ex <- as.numeric(x[2,1:ncol(x)])
Plot a histogram:
>hist(ex)

The above plot is just a basic histogram. Let's add some parameters:
•br=20, #divide the data into 20 bars
•col="blue", #fill in blue color
•main="Histogram of Expression", #title of the histogram
•xlab="Expression", #x axis description
•ylab="Frequency", #y axis description
•freq=TRUE, #y axis is the frequency of each interval
Here is the command:
>hist(ex,br=14,col="blue",xlab="Expression",ylab="Frequency",
+freq=TRUE,main="Histogram of Expression")

To add a density line into the histogram, we need to change two parameters:
•freq=FALSE, #y axis is the density value of each interval
•ylab="Density", #y axis description as Density
Here is the command:
>hist(ex,br=14,col="blue",xlab="Expression",ylab="Density",freq=FALSE,
+main="Histogram of Expression")
>lines(density(ex),col="red")

We can write the plot into a file:
>png("histogram3.png",400,300)
>hist(ex,br=14,col="blue",xlab="Expression",ylab="Density",
+freq=FALSE,main="Histogram of Expression")
>lines(density(ex),col="red")
>graphics.off()
List of hist(...) parameters:
I() Function
I() function changes the class of an object to indicate that it should be treated ‘as is’.
• In function data.frame. Protecting an object by enclosing it in I() in a call to data.frame inhibits the conversion of character vectors to factors and the dropping of names, and ensures that matrices are inserted as single columns. I can also be used to protect objects which are to be added to a data frame, or converted to a data frame via as.data.frame.
It achieves this by prepending the class "AsIs" to the object's classes. Class "AsIs" has a few of its own methods, including for [, as.data.frame, print and format.
• In function formula. There it is used to inhibit the interpretation of operators such as "+", "-", "*" and "^" as formula operators, so they are used as arithmetical operators. This is interpreted as a symbol by terms.formula.
iconv Function
iconv() function uses system facilities to convert a character vector between encodings: the ‘i’ stands for ‘internationalization’.
iconv(x, from = "", to = "", sub = NA, mark = TRUE)
iconvlist()
x: character vector
from: character string describing the current encoding
to: character string describing the target encoding
sub: character string. If not NA it is used to replace any non-convertible bytes in the input. (This would normally be a single character, but can be more.) If "byte", the indication is "
" with the hex code of the byte
mark: logical, for expert use. Should encodings be marked?
icuSetCollate Function
icuSetCollate() function Controls the way collation is done by ICU (an optional part of the R build).
identical Function
identical() function is the safe and reliable way to test whether two objects are exactly equal.
identical(x, y, num.eq = TRUE, single.NA = TRUE,
attrib.as.set = TRUE)
x,y: R object
num.eq: logical indicating if (double and complex non-NA) numbers should be compared using == (‘equal’), or by bitwise comparison. The latter (non-default) differentiates between -0 and +0
single.NA: logical indicating if there is conceptually just one numeric NA and one NaN; single.NA = FALSE differentiates bit patterns
attrib.as.set: logical indicating if attributes of x and y should be treated as unordered tagged pairlists (“sets”); this currently also applies to slots of S4 objects. It may well be too strict to set attrib.as.set = FALSE
> identical(1,1)
[1] TRUE
> identical(1,2/2)
[1] TRUE
> identical(1,"1")
[1] FALSE
> identical(1/0,Inf)
[1] TRUE
> identical(1, as.integer(1))
[1] FALSE
> identical(0,-0)
[1] TRUE
> identical(NaN,-NaN)
[1] TRUE
identity Function
identity() function prints a variable.
identity(x)
x: R object
> identity(BOD)
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
> x <- 5
> identity(x)
[1] 5
IF Else Statement
Syntax: if (condition) {...} else {...}. && and || can be used in the condition. If else statement can be nested.
Let's create a vector containing number 1-10:
>samples <- c(rep(1:10))
>samples
[1] 1 2 3 4 5 6 7 8 9 10
Print out those sample numbers that are even using if else statement:
>for (thissample in samples)
+{
+ if (thissample %% 2 != 0) next
+ else print(thissample)
+}
[1] 2
[1] 4
[1] 6
[1] 8
[1] 10
The
ifelse function is a vectorized version of if else statement. It's syntax is
ifelse(condition,v1,v2). if contidion is true, return v1, otherwise v2.
If we want all samples with number >6 be number 2, and those not be number 1, just:
>ret<-ifelse(samples>6,2,1)
>ret
[1] 1 1 1 1 1 1 2 2 2 2
integer Function
integer() function creates or tests for objects of type integer.
integer(length = 0)
as.integer(x, ...)
is.integer(x)
length: length of the integer vector created
x: R object to be tested
...
> integer(length=3)
[1] 0 0 0
> x <- 3
> is.integer(x)
[1] FALSE
> x <- as.integer(3)
> is.integer(x)
[1] TRUE
interaction Function
interaction() function computes a factor which represents the interaction of the given factors. The result of interaction is always unordered.
interaction(..., drop = FALSE, sep = ".", lex.order = FALSE)
...: the factors for which interaction is to be computed, or a single list giving those factors
drop: if drop is TRUE, unused factor levels are dropped from the result. The default is to retain all factor levels
sep: string to construct the new level labels by joining the constituent ones
lex.order: logical indicating if the order of factor concatenation should be lexically ordered
> x <- gl(2,4)
> y <- gl(2,2)
> x
[1] 1 1 1 1 2 2 2 2
Levels: 1 2
> y
[1] 1 1 2 2
Levels: 1 2
> interaction(x,y)
[1] 1.1 1.1 1.2 1.2 2.1 2.1 2.2 2.2
Levels: 1.1 2.1 1.2 2.2
intersect Function
intersect() function performs intersection on two vectors.
union(x, y)
intersect(x, y)
setdiff(x, y)
setequal(x, y)
is.element(el, set)
x,y,el,set: vectors
> x <- c(1:5)
> x
[1] 1 2 3 4 5
> y <- c(3:8)
> y
[1] 3 4 5 6 7 8
> union(x,y)
[1] 1 2 3 4 5 6 7 8
> intersect(x,y)
[1] 3 4 5
> setdiff(x,y)
[1] 1 2
> setdiff(y,x)
[1] 6 7 8
> setequal(x,y)
[1] FALSE
> is.element(x,y)
[1] FALSE FALSE TRUE TRUE TRUE
intToBits Function
intToBits() function returns a raw vector of 32 times the length of an integer vector with entries 0 and 1.
intToBits(x)
x: Integer
...
> intToBits(1)
[1] 01 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00
00 00 00 00 00 00 00
[26] 00 00 00 00 00 00 00
> intToBits(0)
[1] 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00
00 00 00 00 00 00 00
[26] 00 00 00 00 00 00 00
> intToBits(2)
[1] 00 01 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00
00 00 00 00 00 00 00
[26] 00 00 00 00 00 00 00
> intToBits(3)
[1] 01 01 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00 00
00 00 00 00 00 00 00
[26] 00 00 00 00 00 00 00
intToUtf8 Function
intToUtf8() function converts integer to UTF8.
utf8ToInt(x)
intToUtf8(x, multiple=FALSE)
x: object to be converted
multiple: logical: should the conversion be to a single character string or multiple individual characters?
> intToUtf8(3)
[1] "\003"
> intToUtf8(43)
[1] "+"
> intToUtf8(430)
[1] "Ʈ"
isSymmetric Function
isSymmetric() function tests whether matrix is symmetric or not.
isSymmetric(object, ...)
isSymmetric(object, tol = 100 * .Machine$double.eps, ...)
object: matrix
tol: numeric scalar >= 0
...
> x <- matrix(1:9,3,3)
> x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> isSymmetric(x)
[1] FALSE
> x <- diag(3)
> isSymmetric(x)
[1] TRUE
> x
[,1] [,2] [,3]
[1,] 1 0 0
[2,] 0 1 0
[3,] 0 0 1
isTRUE Function
isTRUE() function tests whether value or express is TRUE or not.
! x
x & y
x && y
x | y
x || y
xor(x, y)
isTRUE(x)
x,y: logical or number-like vectors
...
> isTRUE(1)
[1] FALSE
> isTRUE(0)
[1] FALSE
> isTRUE(1>0)
[1] TRUE
> isTRUE(TRUE)
[1] TRUE
jitter Function
jitter() function adds a small amount of noise to a numeric vector.
jitter(x, factor=1, amount = NULL)
•
x: numeric vector
•
factor: numeric
•
amount: numeric; if positive, used as amount, otherwise, if = 0 the default is factor * z/50. Default (NULL): factor * d/5 where d is about the smallest difference between x values
> jitter(3)
[1] 3.018772
> jitter(3)
[1] 2.987
> jitter(3)
[1] 3.003597
> jitter(3,factor=1000)
[1] -18.7201
> jitter(3,factor=1000)
[1] -47.10491
> jitter(3,factor=1000)
[1] 61.86195
> jitter(3,factor=10,amount=1)
[1] 3.642866
> jitter(rep(4,5))
[1] 4.075898 4.003890 3.934521 3.979925 3.940923
> jitter(rep(2,5))
[1] 2.010194 2.019983 1.997563 2.030967 1.999279
> jitter(rep(0,5))
[1] -0.010258611 -0.003132073 0.017139330 -0.001881735 -0.009311234
kappa Function
kappa() function computes by default (an estimate of) the 2-norm condition number of a matrix or of the R matrix of a QR decomposition, perhaps of a linear fit. The 2-norm condition number can be shown to be the ratio of the largest to the smallest non-zero singular value of the matrix.
The condition number of a regular (square) matrix is the product of the norm of the matrix and the norm of its inverse (or pseudo-inverse), and hence depends on the kind of matrix-norm.
rcond() computes an approximation of the reciprocal condition number.
kappa(z, ...)
kappa(z, exact = FALSE,
norm = NULL, method = c("qr", "direct"), ...)
kappa(z, ...)
kappa(z, ...)
kappa.tri(z, exact = FALSE, LINPACK = TRUE, norm=NULL, ...)
rcond(x, norm = c("O","I","1"), triangular = FALSE, ...)
z,x: A matrix or a the result of qr or a fit from a class inheriting from "lm"
exact: logical. Should the result be exact?
norm: character string, specifying the matrix norm with respect to which the condition number is to be computed, see also norm. For rcond, the default is "O", meaning the One- or 1-norm. The (currently only) other possible value is "I" for the infinity norm
method: character string, specifying the method to be used; "qr" is default for back-compatibility, mainly
triangular: logical. If true, the matrix used is just the lower triangular part of z
LINPACK: logical. If true and z is not complex, the Linpack routine dtrco() is called; otherwise the relevant Lapack routine is
...
> x <- matrix(1:9,3,3)
> kappa(x)
[1] 3.893583e+16
> x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> x <- cbind(2,3:9)
> x
[,1] [,2]
[1,] 2 3
[2,] 2 4
[3,] 2 5
[4,] 2 6
[5,] 2 7
[6,] 2 8
[7,] 2 9
> kappa(x)
[1] 13.6
kronecker Function
kronecker() function computes the generalised kronecker product of two arrays, X and Y. kronecker(X, Y) returns an array A with dimensions dim(X) * dim(Y).
kronecker(X, Y, FUN = "*", make.dimnames = FALSE, ...)
X %x% Y
X: vector, array
Y: vector, array
FUN: function
make.dimnames: dimnames that are the product of the dimnames of X and Y
...
> x <- matrix(1:9,3,3)
> x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> kronecker(2,x)
[,1] [,2] [,3]
[1,] 2 8 14
[2,] 4 10 16
[3,] 6 12 18
> kronecker(5,x)
[,1] [,2] [,3]
[1,] 5 20 35
[2,] 10 25 40
[3,] 15 30 45
> kronecker(diag(3,2),x)
[,1] [,2] [,3] [,4] [,5] [,6]
[1,] 3 12 21 0 0 0
[2,] 6 15 24 0 0 0
[3,] 9 18 27 0 0 0
[4,] 0 0 0 3 12 21
[5,] 0 0 0 6 15 24
[6,] 0 0 0 9 18 27
l10n_info Function
l10n_info() function reports on localization information.
l10n_info()
There are four components:
MBCS: Multi-byte character set in use or not
UTF-8: UTF-8 locale, TRUE or FALSE
Latin-1: Latin-1 locale, TRUE or FALSE
codepage: Codepage value
>l10n_info()
$MBCS
[1] FALSE
$`UTF-8`
[1] FALSE
$`Latin-1`
[1] TRUE
$codepage
[1] 1252
labels Function
labels() function finds a suitable set of labels from an object for use in
printing or plotting.
labels(object, ...)
> x <- 3
> labels(x)
[1] "1"
> x <- c(3,4,5,9)
> labels(x)
[1] "1" "2" "3" "4"
lapply Function
lapply() function applies a function to a data frame.
lapply(x,func, ...)
•
x: array
•
func: the function
...
>BOD #R built-in dataset, Biochemical Oxygen Demand
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
Use
lapply() to sum up all rows, return is a list:
> lapply(BOD,sum)
$Time
[1] 22
$demand
[1] 89
> lapply(BOD,mean)
$Time
[1] 3.666667
$demand
[1] 14.83333
> lapply(BOD,function(x) x*10)
$Time
[1] 10 20 30 40 50 70
$demand
[1] 83 103 190 160 156 198
Add Legends to Plot
legend() function adds a legend box to plot. It's expression is:
legend(x, y = NULL, legend, fill = NULL, col = par("col"),
border = "black", lty, lwd, pch,
angle = 45, density = NULL, bty = "o", bg = par("bg"),
box.lwd = par("lwd"), box.lty = par("lty"), box.col = par("fg"),
pt.bg = NA, cex = 1, pt.cex = cex, pt.lwd = lwd,
xjust = 0, yjust = 1, x.intersp = 1, y.intersp = 1,
adj = c(0, 0.5), text.width = NULL, text.col = par("col"),
text.font = NULL, merge = do.lines && has.pch, trace = FALSE,
plot = TRUE, ncol = 1, horiz = FALSE, title = NULL,
inset = 0, xpd, title.col = text.col, title.adj = 0.5,
seg.len = 2)
x,y:The x and y co-ordinates of the legend
legend:a character vector
fill:Fill the legend box with color
col:Color of the legend content
border:Border color (when legend box is filled)
lty,lwd:Line types and widths of the legend
pch:The plotting symbols appearing in the legend
...
Suppose we have a group of data from some samples, and have a plot:
>x <- c(1.2,3.4,1.3,-2.1,5.6,2.3,3.2,2.4,2.1,1.8,1.7,2.2)
>y <- c(2.4,5.7,2.0,-3,13,5,6.2,4.8,4.2,3.5,3.7,5.2)
>plot(x,y,cex=.8,pch=1,xlab="x",ylab="y",col="black")

Let's add another a group of control data to the plot:
>x2 <- c(4.1,1.1,-2.3,-0.2,-1.2,2.3)
>y2 <- c(2.3,4.2,1.2,2.1,-2,4.3)
>points(x2,y2,cex=.8,pch=3,col="blue")

Then we add a legend to the plot:
>legend(x=-2,y=12,c("sample","control"),cex=.8,
col=c("black","blue"),pch=c(1,3))


See
Scatter Plot for how to produce a legend beside the main plot.
length Function
length() function gets or sets the length of a vector (list) or other objects.
Get vector length:
>x <- c(1,2,5,4,6,1,22,1)
>length(x)
[1] 8
Set vector length:
>length(x) <- 4
>x
[1] 1 2 5 4
Get the length of a list:
>y <- list(batch=3,label="Lung Cancer Patients", subtype=c("A","B","C"))
>y
$batch
[1] 3
$label
[1] "Lung Cancer Patients"
$subtype
[1] "A" "B" "C"
>is.list(y)
[1] TURE
>length(y)
[1] 3
If the parameter is a matrix or dataframe, it returns the number of variables:
>length(BOD)
[1] 2
>BOD
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
When resets a list or matrix, if the list is shortened, extra values will be discarded,
if the list is lengthened,
NAs (or
nul) is added to the list.
> length(BOD) <- 1
> BOD
$Time
[1] 1 2 3 4 5 7
> length(BOD) <- 3
> BOD
$Time
[1] 1 2 3 4 5 7
$demand
[1] 8.3 10.3 19.0 16.0 15.6 19.8
[[3]]
NULL
length() function can be used for all R objects. For an environment it returns the object
number in it. NULL returns 0. Most other objects return length 1.
levels Function
levels() function gets or sets the levels abbribute of a variable.
nlevels() function returns the number of levels of a variable.
levels(x)
levels(x) <- value
nlevels(x)
x: R object
value: for assignment to levels attribute
> x <- 3
> levels(x)
NULL
> x <- c(3,4,5,9)
> levels(x)
NULL
> x <- gl(2,4,5)
> x
[1] 1 1 1 1 2
Levels: 1 2
> levels(x)
[1] "1" "2"
> levels(x) <- c("sample","control")
> levels(x)
[1] "sample" "control"
> x
[1] sample sample sample sample control
Levels: sample control
> x <- gl(2,4,5)
> x
[1] 1 1 1 1 2
Levels: 1 2
> nlevels(x)
[1] 2
library Function
library() and require() function load add-on packages.
library(package, help, pos = 2, lib.loc = NULL,
character.only = FALSE, logical.return = FALSE,
warn.conflicts = TRUE, quietly = FALSE,
keep.source = getOption("keep.source.pkgs"),
verbose = getOption("verbose"))
require(package, lib.loc = NULL, quietly = FALSE,
warn.conflicts = TRUE,
keep.source = getOption("keep.source.pkgs"),
character.only = FALSE, save = FALSE)
.First.lib(libname, pkgname)
.Last.lib(libpath)
package, help: package name
pos: the position on the search list at which to attach the loaded package
lib.loc: a character vector describing the location of R library trees to search through, or NULL. The default value of NULL corresponds to all libraries currently known to .libPaths(). Non-existent library trees are silently ignored
character.only: logical indicating whether package or help can be assumed to be character strings
logical.return: logical. If it is TRUE, FALSE or TRUE is returned to indicate success
warn.conflicts: logical. If TRUE, warnings are printed about conflicts from attaching the new package, unless that package contains an object .conflicts.OK. A conflict is a function masking a function, or a non-function masking a non-function
keep.source: logical. If TRUE, functions ‘keep their source’ including comments, see argument keep.source to options. This applies only to the named package, and not to any packages or name spaces which might be loaded to satisfy dependencies or imports
verbose: a logical. If TRUE, additional diagnostics are printed
quietly: a logical. If TRUE, no message confirming package loading is printed, and most often, no errors/warnings are printed if package loading fails
save: For back-compatibility: only FALSE is allowed
libname: a character string giving the library directory where the package was found
pkgname: a character string giving the name of the package
libpath: package path
Draw Lines
abline() function adds a line to plot. It's expression is:
abline(a = NULL, b = NULL, h = NULL, v = NULL, reg = NULL,
coef = NULL, untf = FALSE, ...)
a,b:Intercept and slope
h:for horizontal line
v:for vertical line
...
First let's make a plot:
>x <- c(1.2,3.4,1.3,-2.1,5.6,2.3,3.2,2.4,2.1,1.8,1.7,2.2)
>y <- c(2.4,5.7,2.0,-3,13,5,6.2,4.8,4.2,3.5,3.7,5.2)
>plot(x,y,cex=.8,pch=1,xlab="x",ylab="y",col="black")
>x2 <- c(4.1,1.1,-2.3,-0.2,-1.2,2.3)
>y2 <- c(2.3,4.2,1.2,2.1,-2,4.3)
>points(x2,y2,cex=.8,pch=3,col="blue")

Let's add a red horizontal line at y=4 to the plot:
>abline(h=4,col="red")

Let's add a green vertical line at x=0 to the plot:
>abline(v=0,col="green")

Let's add a blue line with intercept 2 and slope 2 to the plot:
>abline(a=2,b=2,col="blue")

lty= and
lwd= control the line type and line width.
There are 6 line types:

The line width can be a >0 number, for example, lwd from 1 - 8 as follows:

List Data Type
R list data type refers to an object consisting of an ordered collection of elements.
The elements may be of different mode or type.
Let's create a list containing numeric, character and vector data types:
>x <- list(batch=3,label="Lung Cancer Patients", subtype=c("A","B","C"))
>x
$batch
[1] 3
$label
[1] "Lung Cancer Patients"
$subtype
[1] "A" "B" "C"
>is.list(x)
[1] TRUE
The elements of
list data type are indexed by numbers. e.g. x[[1]] refers to 3 ...
>x[[1]]
[1] 3
x[[2]]
[1] "Lung Cancer Patients"
x[[3]]
[1] "A" "B" "C"
x[[3]][2]
[1] "B"
The elements of
list can also be accessed by their names.
>x$subtype
[1] "A" "B" "C"
>x[["subtype"]]
[1] "A" "B" "C"
The statement
length() calculate the total elements number of a
list.
>length(x)
[1] 3
Function
c() can be used for concatenating two or more lists.
>y <- list(operator="Mary",location="New York")
>z <- list(cost=1000.24,urgent="yes")
>final_list <- c(x,y,z)
>final_list
$batch
[1] 3
$label
[1] "Lung Cancer Patients"
$subtype
[1] "A" "B" "C"
$operator
[1] "Mary"
$location
[1] "New York"
$cost
[1] 1000.24
$urgent
[1] "yes"
List to data frame:
as.data.frame() can coerce a list into a data frame, providing that the components of the list
conforms to the restrictions of a data frame.
>y <- as.data.frame(x)
>y
batch label subtype
1 3 Lung Cancer Patients A
2 3 Lung Cancer Patients B
3 3 Lung Cancer Patients C
List to matrix:
as.matrix() can coerce a list into a matrix, providing that the components of the list
conforms to the restrictions of a matrix.
>y <- as.matrix(x)
>y
[,1]
batch 3
label "Lung Cancer Patients"
subtype Character,3
list2env Function
list2env() function creates an environment containing all list components as objects, or “multi-assign” from x into a pre-existing environment.
list2env(x, envir = NULL, parent = parent.frame(),
hash = (length(x) > 100), size = max(29L, length(x)))
x: list
envir: environment
...
>x <- list(batch=3,label="Lung Cancer Patients", subtype=c("A","B","C"))
>x
$batch
[1] 3
$label
[1] "Lung Cancer Patients"
$subtype
[1] "A" "B" "C"
> e <- list2env(x)
> ls(e)
[1] "batch" "label" "subtype"
load Function
load() function reloads datasets written with the function save.
load(file, envir = parent.frame())
file: binary-mode file
envir: environment for the data to be loaded
> x <- 3
> save(list=ls(all=TRUE),file="tp.RData")
> rm(x)
> load("tp.RData")
> ls()
[1] "e" "x"
> x
[1] 3
log Function
log() function computes natural logarithms (Ln) for a number or vector. log10
computes common logarithms (Lg).log2
computes binary logarithms (Log2). log(x,b) computes logarithms with base b.
>log(5) #ln5
[1] 1.609438
>log10(5) #lg5
[1] 0.69897
>log2(5) #log25
[1] 2.321928
>log(9,base=3) #log39 = 2
[1] 2
Note base is the second parameter.
Let's try vector:
>x <- rep(1:12)
>x
[1] 1 2 3 4 5 6 7 8 9 10 11 12
>log(x)
[1] 0.0000000 0.6931472 1.0986123 1.3862944 1.6094379 1.7917595 1.9459101
[8] 2.0794415 2.1972246 2.3025851 2.3978953 2.4849066
>log(x,6)
[1] 0.0000000 0.3868528 0.6131472 0.7737056 0.8982444 1.0000000 1.0860331
[8] 1.1605584 1.2262944 1.2850972 1.3382908 1.3868528
log10 Function
log10() function computes base 10 logarithm.
log10(x)
x: numeric vector
> log10(100)
[1] 2
> x <- c(100,1000, 10000)
> log10(x)
[1] 2 3 4
log1p Function
log1p(x) function computes log(x+1) accurately.
log1p(x)
x: numeric vector
> log1p(0)
[1] 0
> log1p(1)
[1] 0.6931472
> log1p(-0.1)
[1] -0.1053605
> log1p(9)
[1] 2.302585
> log1p(c(1,0,9))
[1] 0.6931472 0.0000000 2.3025851
log2 Function
log2() function computes binary (base 2) logarithm.
log2(x)
x: numeric vector
> log2(1)
[1] 0
> log2(2)
[1] 1
> log2(8)
[1] 3
> x <- c(1,2,8)
> log2(x)
[1] 0 1 3
match Function
Match() function returns a vector of the positions of (first) matches of vector 1 in vector 2.
If the element of vector 1 is not exist in vector 2, NA is returned.
match(v1, v2, nomatch = NA_integer_, incomparables = NULL)
v1 %in% v2
v1: vector
v2: vector
nomatch: the value to be returned in the case when no match is found
incomparables: a vector of values that cannot be matched. Any value in x matching a value in this vector is assigned the nomatch value. For historical reasons, FALSE is equivalent to NULL
v1 %in% v2 searches all elements of vector v1 in vector v2.
if the elements exists in v2, return TRUE, otherwise FALSE.
> v1 <- c("a","b","c","d")
> v2 <- c("g","x","d","e","f","a","c")
> x <- match(v1,v2)
> x
[1] 6 NA 7 3
> v1 %in% v2
[1] TRUE FALSE TRUE TRUE
> x <- match(v1,v2,nomatch=-1)
> x
[1] 6 -1 7 3
max min Function
max() function computes the maximun value of a vector.
min() function computes the minimum value of a vector.
max(x,na.rm=FALSE)
min(x,na.rm=FALSE)
•
x: number vector
•
na.rm: whether NA should be removed, if not, NA will be returned
...
> x <- c(1,2.3,2,3,4,8,12,43,-4,-1)
> max(x)
[1] 43
> min(x)
[1] -4
Missing value affect the results:
> y<- c(x,NA)
> y
[1] 1.0 2.3 2.0 3.0 4.0 8.0 12.0 43.0 -4.0 -1.0 NA
> max(y)
[1] NA
> min(y)
[1] NA
After define na.rm=TRUE, result is meaningful:
> max(y,na.rm=TRUE)
[1] 43
Compare more than 1 vectors:
> x2 <- c(-100,-43,0,3,1,-3)
> min(x,x2)
[1] -100
mean Function
mean() function calculates the arithmetic mean.
mean(x, trim = 0, na.rm = FALSE, ...)
x: numeric vector
trim: trim off a fraction at each end of the vector, default is 5%
na.rm: whether NA should be removed, if not, NA will be returned
...
>x <- c(1,2.3,2,3,4,8,12,43,-4,-1)
>mean(x)
[1] 7.03
Missing value affect the results:
>y<- c(x,NA)
>y
[1] 1.0 2.3 2.0 3.0 4.0 8.0 12.0 43.0 -4.0 -1.0 NA
>mean(y)
[1] NA
After define na.rm=TRUE, result is meaningful:
>mean(y,na.rm=TRUE)
[1] 43
Trim at each end:
>z <- c(rep(1:20),-200,400)
>mean(z)
[1] 18.63636
>mean(z,trim=0.5)
[1] 10.5
Memory
Memory used in R can be controlled by command line options.
R --min-vsize=vl --max-vsize=vu --min-nsize=nl --max-nsize=nu \
--max-ppsize=N
mem.limits(nsize = NA, vsize = NA)
vl,vu,vsize: Heap memory in bytes
nl,nu,nsize: Number of cons cells
N: Number of nested PROTECT calls
message Function
message() function generates diagnostic message from its arguments.
message(..., domain = NULL, appendLF = TRUE)
suppressMessages(expr)
packageStartupMessage(..., domain = NULL, appendLF = TRUE)
suppressPackageStartupMessages(expr)
.makeMessage(..., domain = NULL, appendLF = FALSE)
...: zero or more objects which can be coerced to character (and which are pasted together with no separator) or (for message only) a single condition object
appendLF: logical: should messages given as a character string have a newline appended?
expr: expression to evaluate
> message("r tutorial")
r tutorial
> message("r tutorial"," ","message")
r tutorial message
missing Function
missing() function tests whether a value was specified as an argument to a function.
missing(x)
x: the argument to be tested
...
myplot <- function(x,y) {
if(missing(y)) {
y <- x
x <- 1:length(y)
}
plot(x,y)
}
mode Function
mode() function gets or sets the type or storage mode of an object.
mode(x)
mode(x) <- value
storage.mode(x)
storage.mode(x) <- value
x: R object
value: character string giving the desired mode or ‘storage mode’ (type) of the object
> x <- 3
> mode(x)
[1] "numeric"
> mode(x) <- "character"
> mode(x)
[1] "character"
name Function
name() function refer to R objects by name (rather than the value of the object, if any, bound to that name).
as.name and as.symbol are identical: they attempt to coerce the argument to a name.
is.symbol and the identical is.name return TRUE or FALSE depending on whether the argument is a name or not.
as.symbol(x)
is.symbol(x)
as.name(x)
is.name(x)
x: R object to be tested
> x <- "sample"
> is.name(x)
[1] FALSE
> x <- as.name("sample")
> is.name(x)
[1] TRUE
> mode(x)
[1] "name"
> typeof(x)
[1] "symbol"
names Function
names() function gets or sets the names of an object.
names(x)
names(x) <- value
x: R object
value: to be assigned to the x, with the same length as x, or NULL
...
> BOD
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
> mode(BOD)
[1] "list"
> names(BOD)
[1] "Time" "demand"
> x <- c(5,7,3)
> names(x) <- c("red","greed","blue")
> names(x)
[1] "red" "greed" "blue"
nargs Function
nargs() function returns the number of arguments supplied to a function, including
positional arguments left blank.
> f <- function(x,y,z=FALSE,...) {nargs();}
> f()
[1] 0
> f(1,2)
[1] 2
nchar Function
nchar() function determines the size of each elements of an character vector. nzchar() tests whether
elements of a character vector are non-empty strings.
nchar(x, type = "chars", allowNA = FALSE)
nzchar(x)
x: character vector
type: bytes, chars or width
allowNA: logical: should NA be returned for invalid multibyte strings or "bytes"-encoded strings (rather than throwing an error)?
> x <- c("red","greed","blue")
> nchar(x)
[1] 3 5 4
> nzchar(x)
[1] TRUE TRUE TRUE
> x <- "red"
> nchar(x)
[1] 3
ncol nrow Function
ncol() function returns the number of columns of a matrix.
nrow() function returns the number of rows of a matrix.
nrow(x)
ncol(x)
NCOL(x)
NROW(x)
x: matrix, vector, array or data frame
> BOD
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
> ncol(BOD)
[1] 2
> nrow(BOD)
[1] 6
> NCOL(BOD)
[1] 2
> NROW(BOD)
[1] 6
noquote Function
noquote() function prints out strings without quotes.
noquote(x)
x: character vector
> letters
[1] "a" "b" "c" "d" "e" "f" "g" "h" "i" "j" "k" "l" "m"
"n" "o" "p" "q" "r" "s"
[20] "t" "u" "v" "w" "x" "y" "z"
> noquote(letters)
[1] a b c d e f g h i j k l m n o p q r s t u v w x y z
> x <- "r tutor\"ial"
> x
[1] "r tutor\"ial"
> noquote(x)
[1] r tutor"ial
norm Function
norm() function computes a matrix norm, by default using Lapack.
norm(x, type = c("O", "I", "F", "M"))
x: numeric matrix
type: the type of matrix norm to be computed. O, o, 1 is the one norm, maximum absolute column sum;
I, i is the infinity norm, maximum absolute row sum; F, f is the Frobenius norm, the Euclidean norm;
M, m is the maximum modulus of all the elements.
> x <- matrix(1:12,3,4)
> x
[,1] [,2] [,3] [,4]
[1,] 1 4 7 10
[2,] 2 5 8 11
[3,] 3 6 9 12
> norm(x)
[1] 33
> norm(x,"I")
[1] 30
> norm(x,"M")
[1] 12
Normality Test
shapiro.test() function performs normality test of a data set with hypothesis that it's normally distributed.
shapiro.test(x)
x: numeric data set
...
Let's generate 100 random number near the range of 0, and to see whether they are normally distributed:
> x <- rnorm(100, mean=0)
> shapiro.test(x)
Shapiro-Wilk normality test
data: x
W = 0.9879, p-value = 0.5011
Since the p-value is > 0.05, it is accepted the dataset is normally distributed.
Let's check the CO2 dataset, Carbon Dioxide Uptake in Grass Plants, to
see whether the CO2 uptake is normally distributed.
> CO2
Plant Type Treatment conc uptake
1 Qn1 Quebec nonchilled 95 16.0
2 Qn1 Quebec nonchilled 175 30.4
3 Qn1 Quebec nonchilled 250 34.8
4 Qn1 Quebec nonchilled 350 37.2
5 Qn1 Quebec nonchilled 500 35.3
6 Qn1 Quebec nonchilled 675 39.2
7 Qn1 Quebec nonchilled 1000 39.7
8 Qn2 Quebec nonchilled 95 13.6
9 Qn2 Quebec nonchilled 175 27.3
10 Qn2 Quebec nonchilled 250 37.1
11 Qn2 Quebec nonchilled 350 41.8
12 Qn2 Quebec nonchilled 500 40.6
13 Qn2 Quebec nonchilled 675 41.4
14 Qn2 Quebec nonchilled 1000 44.3
15 Qn3 Quebec nonchilled 95 16.2
16 Qn3 Quebec nonchilled 175 32.4
17 Qn3 Quebec nonchilled 250 40.3
18 Qn3 Quebec nonchilled 350 42.1
19 Qn3 Quebec nonchilled 500 42.9
20 Qn3 Quebec nonchilled 675 43.9
21 Qn3 Quebec nonchilled 1000 45.5
22 Qc1 Quebec chilled 95 14.2
23 Qc1 Quebec chilled 175 24.1
24 Qc1 Quebec chilled 250 30.3
25 Qc1 Quebec chilled 350 34.6
26 Qc1 Quebec chilled 500 32.5
27 Qc1 Quebec chilled 675 35.4
28 Qc1 Quebec chilled 1000 38.7
29 Qc2 Quebec chilled 95 9.3
30 Qc2 Quebec chilled 175 27.3
31 Qc2 Quebec chilled 250 35.0
32 Qc2 Quebec chilled 350 38.8
33 Qc2 Quebec chilled 500 38.6
34 Qc2 Quebec chilled 675 37.5
35 Qc2 Quebec chilled 1000 42.4
36 Qc3 Quebec chilled 95 15.1
37 Qc3 Quebec chilled 175 21.0
38 Qc3 Quebec chilled 250 38.1
39 Qc3 Quebec chilled 350 34.0
40 Qc3 Quebec chilled 500 38.9
41 Qc3 Quebec chilled 675 39.6
42 Qc3 Quebec chilled 1000 41.4
43 Mn1 Mississippi nonchilled 95 10.6
44 Mn1 Mississippi nonchilled 175 19.2
45 Mn1 Mississippi nonchilled 250 26.2
46 Mn1 Mississippi nonchilled 350 30.0
47 Mn1 Mississippi nonchilled 500 30.9
48 Mn1 Mississippi nonchilled 675 32.4
49 Mn1 Mississippi nonchilled 1000 35.5
50 Mn2 Mississippi nonchilled 95 12.0
51 Mn2 Mississippi nonchilled 175 22.0
52 Mn2 Mississippi nonchilled 250 30.6
53 Mn2 Mississippi nonchilled 350 31.8
54 Mn2 Mississippi nonchilled 500 32.4
55 Mn2 Mississippi nonchilled 675 31.1
56 Mn2 Mississippi nonchilled 1000 31.5
57 Mn3 Mississippi nonchilled 95 11.3
58 Mn3 Mississippi nonchilled 175 19.4
59 Mn3 Mississippi nonchilled 250 25.8
60 Mn3 Mississippi nonchilled 350 27.9
61 Mn3 Mississippi nonchilled 500 28.5
62 Mn3 Mississippi nonchilled 675 28.1
63 Mn3 Mississippi nonchilled 1000 27.8
64 Mc1 Mississippi chilled 95 10.5
65 Mc1 Mississippi chilled 175 14.9
66 Mc1 Mississippi chilled 250 18.1
67 Mc1 Mississippi chilled 350 18.9
68 Mc1 Mississippi chilled 500 19.5
69 Mc1 Mississippi chilled 675 22.2
70 Mc1 Mississippi chilled 1000 21.9
71 Mc2 Mississippi chilled 95 7.7
72 Mc2 Mississippi chilled 175 11.4
73 Mc2 Mississippi chilled 250 12.3
74 Mc2 Mississippi chilled 350 13.0
75 Mc2 Mississippi chilled 500 12.5
76 Mc2 Mississippi chilled 675 13.7
77 Mc2 Mississippi chilled 1000 14.4
78 Mc3 Mississippi chilled 95 10.6
79 Mc3 Mississippi chilled 175 18.0
80 Mc3 Mississippi chilled 250 17.9
81 Mc3 Mississippi chilled 350 17.9
82 Mc3 Mississippi chilled 500 17.9
83 Mc3 Mississippi chilled 675 18.9
84 Mc3 Mississippi chilled 1000 19.9
> y <- CO2[,5]
> shapiro.test(y)
Shapiro-Wilk normality test
data: y
W = 0.941, p-value = 0.0007908
Since the p-value is smaller than 0.05, it's rejected that the CO2 uptake is normally distributed.
normalizePath Function
normalizePath() function Convert file paths to canonical form for the platform, to display them in a user-understandable form and so that relative and absolute paths can be compared.
normalizePath(path, winslash = "\\", mustWork = NA)
path: character vector
winslash: the separator to be used on Windows – ignored elsewhere. Must be one of c("/", "\\")
mustWork: logical: if TRUE then an error is given if the result cannot be determined; if NA then a warning
> path <- getwd()
> path
[1] "C:/program/r"
> normalizePath(path)
[1] "C:\\program\\r"
> p2 <- "../"
> normalizePath(p2)
[1] "C:\\program"
> normalizePath(p2, winslash="/")
[1] "C:/program"
octmode Function
octmode() function converts or prints integers in octal format, with as many digits as are needed to display the largest, using leading zeroes as necessary.
as.octmode(x)
x: R object
> x <- 3
> as.octmode(x)
[1] "3"
> x <- 145
> as.octmode(x)
[1] "221"
open Function
open() function opens a connection.
open(con, mode = "r", blocking = TRUE, ...)
isOpen(con,rw="")
con: connection handle
mode: description of how to open the connection (if it should be opened initially)
blocking: logical.
...
Open modes list:
| mode | description |
| r or rt | read in text mode |
| w or wt | write in text mode |
| a or at | append in text mode |
| rb | read in binary mode |
| wb | write in binary mode |
| ab | append in binary mode |
| r+, or r+b | read and write |
| w+ or w+b | read and write, truncating file initially |
| a+ or a+b | read and append |
Operators
| + | Add, 2 + 3 = 5 |
| - | Subtract, 5 - 2 = 3 |
| * | Multiply, 2 * 3 = 6 |
| / | Divide, 6 / 2 = 3 |
| ^ | Exponent, 2 ^ 3 = 8 |
| %% | Modulus operator, 9%%2 = 1 |
| %/% | Integer division, 9 %/% 2 = 4 |
| < | Less than |
| > | Greater than |
| = | Equal to |
| <= | Less than or equal to |
| >= | Greater than or equal to |
| != | Not equal to |
| ! | Not |
| | | OR |
| & | And |
Define new operators:
Let's define "+" not as add, but as multiply:
>'+' <- function(x,y) x * y
>3 + 5
[1] 15
Let's delete the self defined operator "+":
>rm('+')
>3 + 5
[1] 8
options Function
options() function allows the user to set and examine a variety of global options which affect the way in which R computes and displays its results.
options(...)
getOption(x, default = NULL)
.Options
...: any options can be defined, using name = value or by passing a list of such tagged values. However, only the ones below are used in base R. Further, options('name') == options()['name'], see the example
x: a character string holding an option name
default: if the specified option is not set in the options list, this value is returned. This facilitates retrieving an option and checking whether it is set and setting it separately if not
order Function
order() function sorts a vector, matrix or data frame.
order(x, decreasing = FALSE, na.last = NA, ...)
x: vector
decreasing: decrease or not
na.last: if TRUE, NAs are put at last position, FALSE at first, if NA, remove them (default)
...
Sort Vectors:
>x <- c(1,2.3,2,3,4,8,12,43,-4,-1,NA)
>order(x)
[1] -4.0 -1.0 1.0 2.0 2.3 3.0 4.0 8.0 12.0 43.0
>order(x,decreasing=TRUE)
[1] 43.0 12.0 8.0 4.0 3.0 2.3 2.0 1.0 -1.0 -4.0
>order(x,decreasing=TRUE, na.last=TRUE)
[1] 43.0 12.0 8.0 4.0 3.0 2.3 2.0 1.0 -1.0 -4.0 NA
>order(x,decreasing=TRUE, na.last=FALSE)
[1] NA 43.0 12.0 8.0 4.0 3.0 2.3 2.0 1.0 -1.0 -4.0
Sort Matrix by one column, following is a csv file example.
>x <- read.csv("ordermatrix.csv",header=T,sep=",");
>x <- x[order(x[,4]),];
>x
Order data frame:
>BOD #R built-in dataset, Biochemical Oxygen Demand
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
Sort by "demand" column:
>BOD[with(BOD,order(demand)),]
Time demand
1 1 8.3
2 2 10.3
5 5 15.6
4 4 16.0
3 3 19.0
6 7 19.8
outer Function
outer() function applies a function to two arrays.
outer(x, y, FUN="*", ...)
x %o% y
x,y: arrays
FUN: function to use on the outer products, default is multiply
...
>x <- c(1,2.3,2,3,4,8,12,43)
>y<- c(2,4)
Calculate logarithm value of array x elements using array y as bases:
>outer(x,y,"log")
[,1] [,2]
[1,] 0.000000 0.0000000
[2,] 1.201634 0.6008169
[3,] 1.000000 0.5000000
[4,] 1.584963 0.7924813
[5,] 2.000000 1.0000000
[6,] 3.000000 1.5000000
[7,] 3.584963 1.7924813
[8,] 5.426265 2.7131324
Add array x elements with array y elements:
> outer(x,y,"+")
[,1] [,2]
[1,] 3.0 5.0
[2,] 4.3 6.3
[3,] 4.0 6.0
[4,] 5.0 7.0
[5,] 6.0 8.0
[6,] 10.0 12.0
[7,] 14.0 16.0
[8,] 45.0 47.0
Multiply array x elements with array y elements:
> x %o% y #equal to outer(x,y,"*")
[,1] [,2]
[1,] 2.0 4.0
[2,] 4.6 9.2
[3,] 4.0 8.0
[4,] 6.0 12.0
[5,] 8.0 16.0
[6,] 16.0 32.0
[7,] 24.0 48.0
[8,] 86.0 172.0
Concatenate characters to the array elements:
>z <- c("a","b")
>outer(x,z,"paste")
[,1] [,2]
[1,] "1 a" "1 b"
[2,] "2.3 a" "2.3 b"
[3,] "2 a" "2 b"
[4,] "3 a" "3 b"
[5,] "4 a" "4 b"
[6,] "8 a" "8 b"
[7,] "12 a" "12 b"
[8,] "43 a" "43 b"
parse Function
parse() function returns the parsed but unevaluated expressions in a list.
parse(file = "", n = NULL, text = NULL, prompt = "?", srcfile,
encoding = "unknown")
file: a connection, or a character string giving the name of a file or a URL to read the expressions from. If file is "" and text is missing or NULL then input is taken from the console
n: the maximum number of expressions to parse. If n is NULL or negative or NA the input is parsed in its entirety.
text: character vector. The text to parse. Elements are treated as if they were lines of a file. Other R objects will be coerced to character if possible
prompt: the prompt to print when parsing from the keyboard. NULL means to use R's prompt, getOption("prompt")
srcfile: NULL, or a srcfile object
encoding: encoding to be assumed for input strings. If the value is "latin1" or "UTF-8" it is used to mark character strings as known to be in Latin-1 or UTF-8: it is not used to re-encode the input. To do the latter, specify the encoding as part of the connection con or via options(encoding=)
Paste
Concatenate vectors after converting to character.
Usage:
paste(..., sep = " ", collapse = NULL)
Arguments:
...: |
one or more R objects, to be converted to character vectors. |
sep: |
a character string to separate the terms. |
collapse: |
an optional character string to separate the results. |
Details:
paste converts its arguments (via
as.character) to character strings, and concatenates
them (separating them by the string given by sep). If the
arguments are vectors, they are concatenated term-by-term to give a
character vector result. Vector arguments are recycled as needed,
with zero-length arguments being recycled to "".
Note that paste() coerces NA_character_, the
character missing value, to "NA" which may seem
undesirable, e.g., when pasting two character vectors, or very
desirable, e.g. in paste("the value of p is ", p).
If a value is specified for collapse, the values in the result
are then concatenated into a single string, with the elements being
separated by the value of collapse.
Value:
A character vector of the concatenated values. This will be of length
zero if all the objects are, unless collapse is non-NULL in
which case it is a single empty string.
If any input into an element of the result is in UTF-8 (and none are
declared with encoding "bytes"), that element will be in UTF-8,
otherwise in the current encoding in which case the encoding of the
element is declared if the current locale is either Latin-1 or UTF-8,
at least one of the corresponding inputs (including separators) had a
declared encoding and all inputs were either ASCII or declared.
If an input into an element is declared with encoding "bytes",
no translation will be done of any of the elements and the resulting
element will have encoding "bytes". If collapse is
non-NULL, this applies also to the second, collapsing, phase, but some
translation may have been done in pasting object together in the first
phase.
Plot PCH Symbols Chart
Following is a chart of PCH symbols used in R plot. When the PCH is 21-25, the parameter "col=" and "bg=" should be specified.
PCH can also be in characters, such as "#", "%", "A", "a", and the character will be ploted.

Values pch=26:32 are currently unused, and pch=32:255 give the text symbol in a single-byte locale. In a multi-byte locale such as UTF-8, numeric values of pch greater than or equal to 32 specify a Unicode code point.
If pch is an integer or character NA or an empty character string, the point is omitted from the plot.
Value pch="." is handled specially. It is a rectangle of side 0.01 inch (scaled by cex). In addition, if cex = 1 (the default), each side is at least one pixel (1/72 inch on the pdf, postscript and xfig devices).
pch=0,square
pch=1,circle
pch=2,triangle point up
pch=3,plus
pch=4,cross
pch=5,diamond
pch=6,triangle point down
pch=7,square cross
pch=8,star
pch=9,diamond plus
pch=10,circle plus
pch=11,triangles up and down
pch=12,square plus
pch=13,circle cross
pch=14,square and triangle down
pch=15, filled square blue
pch=16, filled circle blue
pch=17, filled triangle point up blue
pch=18, filled diamond blue
pch=19,solid circle blue
pch=20,bullet (smaller circle)
pch=21, filled circle red
pch=22, filled square red
pch=23, filled diamond red
pch=24, filled triangle point up red
pch=25, filled triangle point down red
By default, pch=1 if not specified, and in black color:
>x <- c(2,1,3,2,5,3.3,1.4);
>y <- c(4,2.7,6,3,8,6,2.2);
>plot(x,y)

cex controls the symbol size in the plot, default is
cex=1,
col controls the color of the symbol border, default is
col="black".
Plot with specified PCH, Color and Size:
>plot(x,y,pch=2,cex=4,col="red")

Pie Chart Plot
pie(...) funtion plot a pie chart. It's usage is:
pie(x, labels = names(x), edges = 200, radius = 0.8,
clockwise = FALSE, init.angle = if(clockwise) 90 else 0,
density = NULL, angle = 45, col = NULL, border = NULL,
lty = NULL, main = NULL, ...)
x: Vector of each pie slice areas
labels: Vector of Pie slice names
edges: Pie circle border
radius: Pie circle radius
clockwise: Data direction, default is not clockwise
...
First let's make a simple pie chart:
>x <- c(3,2,6,8,4)
>Pie(x)

Let's add some annotations, including a title (
main=),
color (
col=), pie slice names (
labels=),
etc:
>pieplot(x,labels=c("Jan","Feb","Mar","Apr","May"),xlab="Month",
+ ylab="Revenue", col=c("tan2","darkslategray3","blue","red","green"),
+ density=c(0,5,20,50,100), main="Soft Revenue")

Plot Function
plot(...) is a generic X Y plotting function. It's usage is:
plot(x, y = NULL, type = "p", xlim = NULL, ylim = NULL,
log = "", main = NULL, sub = NULL, xlab = NULL, ylab = NULL,
ann = par("ann"), axes = TRUE, frame.plot = axes,
panel.first = NULL, panel.last = NULL, asp = NA, ...)
x,y:Vector of coordinates
First let's make a simple plot:
>x <- c(1.2,3.4,1.3,-2.1,5.6,2.3,3.2,2.4,2.1,1.8,1.7,2.2)
>y <- c(2.4,5.7,2.0,-3,13,5,6.2,4.8,4.2,3.5,3.7,5.2)
>plot(x,y)

The
par(...) controls the general layout of the plot. For example,
par(mar = c(5, 4, 2, 1)) defines the bottom margin as 5, left margin 4,
top margin 2 and right margin as 1.
The default type is a point plot (type="p"). The possible types include:
| p: | Points, default |
| l: | Lines |
| b: | Points with line connection |
| c: | Line connections without points |
| o: | Both overplotted |
| h: | Histogram like vertical lines |
| s: | Stair steps |
| S: | Stair steps, another style |
| n: | No plotting |
Let's use less points and plot with line connections. We will
use blue colored line and points, and with axis labels both to X and Y axis as well as a main title of the plot:
>x <- c(-2,-0.3,1.4,2.4,4.5)
>y <- c(5,-0.5,8,2,11)
>par(mar = c(5, 1, 1, 1))
>plot(x,y,type="l",col="blue",xlab="Advertise Change",
+ ylab="Revenue Change", main="Financial Analysis")

Add more data to the plot:
>abline(v=0,col="red") #add a vertical line at x=0
>points(c(1,4),c(9,2),pch=3,col="tan2") #add two points
>x2 <- c(-1.5,1,4)
>y2 <- c(3,2,8)
>lines(x2,y2,col="darkolivegreen3") #add aother group of data
>legend(x=-2.2,y=11,c("advertise","sale"),cex=.8,
+ col=c("blue","darkolivegreen3"),lty=c(1,1)) #add legend

If we want to move the legend out of the main plot area, we need some more work.
First use
layout(...) function to define 2 plots on one layer side by side,
and then we plot the same data on both plots, with the plot on the right side in white color,
thus invisible (just providing the scale), and finally we plot the legend on the second plot.
>x <- c(-2,-0.3,1.4,2.4,4.5)
>y <- c(5,-0.5,8,2,11)
>layout(matrix(c(1,2), nrow = 1), widths = c(0.6, 0.4))
>par(mar = c(5, 4, 2, 1))
>plot(x,y,type="b",col="blue",xlab="Advertise Change",
+ ylab="Revenue Change", main="Financial Analysis")
>abline(v=0,col="red") #add a vertical line at x=0
>points(c(1,4),c(9,2),pch=3,col="tan2") #add two points
>x2 <- c(-1.5,1,4)
>y2 <- c(3,2,8)
>lines(x2,y2,col="darkolivegreen3")
>par(mar = c(5, 0, 2, 1))
>plot(x,y,col="white",axes=FALSE,ann=FALSE)
>legend(x=-2.2,y=11,c("advertise","sale"),cex=.8,
+ col=c("blue","darkolivegreen3"),lty=c(1,1))

pmatch Function
pmatch() function seeks matches for the elements of its first argument among those of its second.
pmatch(v1, v2, nomatch = NA_integer_, duplicates.ok = FALSE)
v1: vector
v2: vector
nomatch: the value to be returned in the case when no match is found
duplicates.ok: should elements be in table be used more than once?
> x <- c("green","red","yellow","blue")
> x
[1] "green" "red" "yellow" "blue"
> pmatch("re",x)
[1] 2
> pmatch("e",x)
[1] NA
> pmatch("ye",x)
[1] 3
> pmatch(c("re","ye"),x)
[1] 2 3
pmax Function
pmax() function returns the parallel maxima vector of
multiple vectors or matrix.
pmax(..., na.rm = FALSE)
...: Numeric or character arguments
na.rm: whether missing values should be removed
> x <- c(3, 26, 122, 6)
> y <- c(43,2,54,8)
> z <- c(9,32,1,9)
> pmax(x,y,z)
[1] 43 32 122 9
pmin Function
pmin() function returns the parallel minima vector of
multiple vectors or matrix.
pmin(..., na.rm = FALSE)
...: Numeric or character arguments
na.rm: whether missing values should be removed
> x <- c(3, 26, 122, 6)
> y <- c(43,2,54,8)
> z <- c(9,32,1,9)
> pmax(x,y,z)
[1] 43 32 122 9
> pmin(x,y,z)
[1] 3 2 1 6
Draw Points
points(...) function adds a group of points to plot. It's usage is:
points(x, y, ...)
x,y:Vector of coordinates
First let's make a scatter plot:
>x <- c(1.2,3.4,1.3,-2.1,5.6,2.3,3.2,2.4,2.1,1.8,1.7,2.2)
>y <- c(2.4,5.7,2.0,-3,13,5,6.2,4.8,4.2,3.5,3.7,5.2)
>plot(x,y,cex=.8,pch=1,xlab="x",ylab="y",col="black")

Add some points to the plot:
>x2 <- c(4.1,1.1,-2.3,-0.2,-1.2,2.3)
>y2 <- c(2.3,4.2,1.2,2.1,-2,4.3)
>points(x2,y2,cex=.8,pch=3,col="blue")

Notice that there is a point almost out of the left border. If the added points are out of the plot border,
they were not be added to the plot. In the example above, the smallest value of x is -2.1, and largest is
5.6, the y value range is -3 < y < 13, so the added points should be inside that range.
The
cex= controls the size of the points,
pch= controls
the point shape, and
col= controls the point color.
Here
is a list of all pch symbols, and
here is a complete chart of R color names.
Let add some points of filled diamond shape, large size, and red color:
>x3 <- c(0,4)
>y3 <- c(10,-0.5)
>points(x3,y3,cex=4,pch=18,col="red")

polyroot Function
polyroot() function finds zero of a real or complex polynomail.
polyroot(z)
z: the vector of polynomial coefficients in increasing order
> x <- c(3, 26, 122, 6)
> y <- c(43,2,54,8)
> z <- c(9,32,1,9)
> polyroot(x)
[1] -0.107074+0.1157025i -0.107074-0.1157025i -20.119185+0.0000000i
> polyroot(y)
[1] 0.0393287+0.886328i 0.0393287-0.886328i -6.8286573+0.000000i
> polyroot(z)
[1] -0.2776397+0.000000i 0.0832643+1.896011i 0.0832643-1.896011i
pretty Function
pretty() function computes a sequence of about n+1 equally spaced ‘round’ values which cover the range of the values in x. The values are chosen so that they are 1, 2 or 5 times a power of 10.
pretty(x, n = 5, min.n = n %/% 3, shrink.sml = 0.75,
high.u.bias = 1.5, u5.bias = .5 + 1.5*high.u.bias,
eps.correct = 0, ...)
x: numeric object
n: number of intervals
min.n: nonnegative integer giving the minimal number of intervals. If min.n == 0, pretty(.) may return a single value
shrink.sml: positive numeric by a which a default scale is shrunk in the case when range(x) is very small (usually 0)
high.u.bias: non-negative numeric, typically > 1. The interval unit is determined as {1,2,5,10} times b, a power of 10. Larger high.u.bias values favor larger units
u5.bias: non-negative numeric multiplier favoring factor 5 over 2. Default and ‘optimal’: u5.bias = .5 + 1.5*high.u.bias
eps.correct: integer code, one of {0,1,2}. If non-0, an epsilon correction is made at the boundaries such that the result boundaries will be outside range(x); in the small case, the correction is only done if eps.correct >=2
...
> pretty(pi)
[1] 2 4
> pretty(7)
[1] 5 10
> pretty(33)
[1] 30 40
> pretty(133)
[1] 120 140
> pretty(1:12)
[1] 0 2 4 6 8 10 12
> pretty(1:15)
[1] 0 2 4 6 8 10 12 14 16
> pretty(1:25)
[1] 0 5 10 15 20 25
proc.time Function
proc.time() function determines how much real and CPU time (in seconds) the currently running R process has already taken.
> t0 <- proc.time()
> for (i in 1:500000) print("1")
> proc.time() - t0
user system elapsed
7.06 0.02 41.05
prod Function
prod() function returns the multiplication results of all the values present in its arguments.
prod(..., na.rm=FALSE)
...: numeric or complex or logical vectors
na.rm: whether missing values be removed or not
...
> prod(4:6) #4 × 5 × 6
[1] 120
> x <- c(3.2,5,4.3)
> prod(x) #3.2 × 5 × 4.3
[1] 68.8
Colors Chart
R has 657 built-in color names. The function colors() will show all of them.
All these color names can be used in plot parameters like col=. The function
col2rgb() can convert all these colors into RGB numbers.
| white |
aliceblue |
antiquewhite |
antiquewhite1 |
| antiquewhite2 |
antiquewhite3 |
antiquewhite4 |
aquamarine |
| aquamarine1 |
aquamarine2 |
aquamarine3 |
aquamarine4 |
| azure |
azure1 |
azure2 |
azure3 |
| azure4 |
beige |
bisque |
bisque1 |
| bisque2 |
bisque3 |
bisque4 |
black |
| blanchedalmond |
blue |
blue1 |
blue2 |
| blue3 |
blue4 |
blueviolet |
brown |
| brown1 |
brown2 |
brown3 |
brown4 |
| burlywood |
burlywood1 |
burlywood2 |
burlywood3 |
| burlywood4 |
cadetblue |
cadetblue1 |
cadetblue2 |
| cadetblue3 |
cadetblue4 |
chartreuse |
chartreuse1 |
| chartreuse2 |
chartreuse3 |
chartreuse4 |
chocolate |
| chocolate1 |
chocolate2 |
chocolate3 |
chocolate4 |
| coral |
coral1 |
coral2 |
coral3 |
| coral4 |
cornflowerblue |
cornsilk |
cornsilk1 |
| cornsilk2 |
cornsilk3 |
cornsilk4 |
cyan |
| cyan1 |
cyan2 |
cyan3 |
cyan4 |
| darkblue |
darkcyan |
darkgoldenrod |
darkgoldenrod1 |
| darkgoldenrod2 |
darkgoldenrod3 |
darkgoldenrod4 |
darkgray |
| darkgreen |
darkgrey |
darkkhaki |
darkmagenta |
| darkolivegreen |
darkolivegreen1 |
darkolivegreen2 |
darkolivegreen3 |
| darkolivegreen4 |
darkorange |
darkorange1 |
darkorange2 |
| darkorange3 |
darkorange4 |
darkorchid |
darkorchid1 |
| darkorchid2 |
darkorchid3 |
darkorchid4 |
darkred |
| darksalmon |
darkseagreen |
darkseagreen1 |
darkseagreen2 |
| darkseagreen3 |
darkseagreen4 |
darkslateblue |
darkslategray |
| darkslategray1 |
darkslategray2 |
darkslategray3 |
darkslategray4 |
| darkslategrey |
darkturquoise |
darkviolet |
deeppink |
| deeppink1 |
deeppink2 |
deeppink3 |
deeppink4 |
| deepskyblue |
deepskyblue1 |
deepskyblue2 |
deepskyblue3 |
| deepskyblue4 |
dimgray |
dimgrey |
dodgerblue |
| dodgerblue1 |
dodgerblue2 |
dodgerblue3 |
dodgerblue4 |
| firebrick |
firebrick1 |
firebrick2 |
firebrick3 |
| firebrick4 |
floralwhite |
forestgreen |
gainsboro |
| ghostwhite |
gold |
gold1 |
gold2 |
| gold3 |
gold4 |
goldenrod |
goldenrod1 |
| goldenrod2 |
goldenrod3 |
goldenrod4 |
gray |
| gray0 |
gray1 |
gray2 |
gray3 |
| gray4 |
gray5 |
gray6 |
gray7 |
| gray8 |
gray9 |
gray10 |
gray11 |
| gray12 |
gray13 |
gray14 |
gray15 |
| gray16 |
gray17 |
gray18 |
gray19 |
| gray20 |
gray21 |
gray22 |
gray23 |
| gray24 |
gray25 |
gray26 |
gray27 |
| gray28 |
gray29 |
gray30 |
gray31 |
| gray32 |
gray33 |
gray34 |
gray35 |
| gray36 |
gray37 |
gray38 |
gray39 |
| gray40 |
gray41 |
gray42 |
gray43 |
| gray44 |
gray45 |
gray46 |
gray47 |
| gray48 |
gray49 |
gray50 |
gray51 |
| gray52 |
gray53 |
gray54 |
gray55 |
| gray56 |
gray57 |
gray58 |
gray59 |
| gray60 |
gray61 |
gray62 |
gray63 |
| gray64 |
gray65 |
gray66 |
gray67 |
| gray68 |
gray69 |
gray70 |
gray71 |
| gray72 |
gray73 |
gray74 |
gray75 |
| gray76 |
gray77 |
gray78 |
gray79 |
| gray80 |
gray81 |
gray82 |
gray83 |
| gray84 |
gray85 |
gray86 |
gray87 |
| gray88 |
gray89 |
gray90 |
gray91 |
| gray92 |
gray93 |
gray94 |
gray95 |
| gray96 |
gray97 |
gray98 |
gray99 |
| gray100 |
green |
green1 |
green2 |
| green3 |
green4 |
greenyellow |
grey |
| grey0 |
grey1 |
grey2 |
grey3 |
| grey4 |
grey5 |
grey6 |
grey7 |
| grey8 |
grey9 |
grey10 |
grey11 |
| grey12 |
grey13 |
grey14 |
grey15 |
| grey16 |
grey17 |
grey18 |
grey19 |
| grey20 |
grey21 |
grey22 |
grey23 |
| grey24 |
grey25 |
grey26 |
grey27 |
| grey28 |
grey29 |
grey30 |
grey31 |
| grey32 |
grey33 |
grey34 |
grey35 |
| grey36 |
grey37 |
grey38 |
grey39 |
| grey40 |
grey41 |
grey42 |
grey43 |
| grey44 |
grey45 |
grey46 |
grey47 |
| grey48 |
grey49 |
grey50 |
grey51 |
| grey52 |
grey53 |
grey54 |
grey55 |
| grey56 |
grey57 |
grey58 |
grey59 |
| grey60 |
grey61 |
grey62 |
grey63 |
| grey64 |
grey65 |
grey66 |
grey67 |
| grey68 |
grey69 |
grey70 |
grey71 |
| grey72 |
grey73 |
grey74 |
grey75 |
| grey76 |
grey77 |
grey78 |
grey79 |
| grey80 |
grey81 |
grey82 |
grey83 |
| grey84 |
grey85 |
grey86 |
grey87 |
| grey88 |
grey89 |
grey90 |
grey91 |
| grey92 |
grey93 |
grey94 |
grey95 |
| grey96 |
grey97 |
grey98 |
grey99 |
| grey100 |
honeydew |
honeydew1 |
honeydew2 |
| honeydew3 |
honeydew4 |
hotpink |
hotpink1 |
| hotpink2 |
hotpink3 |
hotpink4 |
indianred |
| indianred1 |
indianred2 |
indianred3 |
indianred4 |
| ivory |
ivory1 |
ivory2 |
ivory3 |
| ivory4 |
khaki |
khaki1 |
khaki2 |
| khaki3 |
khaki4 |
lavender |
lavenderblush |
| lavenderblush1 |
lavenderblush2 |
lavenderblush3 |
lavenderblush4 |
| lawngreen |
lemonchiffon |
lemonchiffon1 |
lemonchiffon2 |
| lemonchiffon3 |
lemonchiffon4 |
lightblue |
lightblue1 |
| lightblue2 |
lightblue3 |
lightblue4 |
lightcoral |
| lightcyan |
lightcyan1 |
lightcyan2 |
lightcyan3 |
| lightcyan4 |
lightgoldenrod |
lightgoldenrod1 |
lightgoldenrod2 |
| lightgoldenrod3 |
lightgoldenrod4 |
lightgoldenrodyellow |
lightgray |
| lightgreen |
lightgrey |
lightpink |
lightpink1 |
| lightpink2 |
lightpink3 |
lightpink4 |
lightsalmon |
| lightsalmon1 |
lightsalmon2 |
lightsalmon3 |
lightsalmon4 |
| lightseagreen |
lightskyblue |
lightskyblue1 |
lightskyblue2 |
| lightskyblue3 |
lightskyblue4 |
lightslateblue |
lightslategray |
| lightslategrey |
lightsteelblue |
lightsteelblue1 |
lightsteelblue2 |
| lightsteelblue3 |
lightsteelblue4 |
lightyellow |
lightyellow1 |
| lightyellow2 |
lightyellow3 |
lightyellow4 |
limegreen |
| linen |
magenta |
magenta1 |
magenta2 |
| magenta3 |
magenta4 |
maroon |
maroon1 |
| maroon2 |
maroon3 |
maroon4 |
mediumaquamarine |
| mediumblue |
mediumorchid |
mediumorchid1 |
mediumorchid2 |
| mediumorchid3 |
mediumorchid4 |
mediumpurple |
mediumpurple1 |
| mediumpurple2 |
mediumpurple3 |
mediumpurple4 |
mediumseagreen |
| mediumslateblue |
mediumspringgreen |
mediumturquoise |
mediumvioletred |
| midnightblue |
mintcream |
mistyrose |
mistyrose1 |
| mistyrose2 |
mistyrose3 |
mistyrose4 |
moccasin |
| navajowhite |
navajowhite1 |
navajowhite2 |
navajowhite3 |
| navajowhite4 |
navy |
navyblue |
oldlace |
| olivedrab |
olivedrab1 |
olivedrab2 |
olivedrab3 |
| olivedrab4 |
orange |
orange1 |
orange2 |
| orange3 |
orange4 |
orangered |
orangered1 |
| orangered2 |
orangered3 |
orangered4 |
orchid |
| orchid1 |
orchid2 |
orchid3 |
orchid4 |
| palegoldenrod |
palegreen |
palegreen1 |
palegreen2 |
| palegreen3 |
palegreen4 |
paleturquoise |
paleturquoise1 |
| paleturquoise2 |
paleturquoise3 |
paleturquoise4 |
palevioletred |
| palevioletred1 |
palevioletred2 |
palevioletred3 |
palevioletred4 |
| papayawhip |
peachpuff |
peachpuff1 |
peachpuff2 |
| peachpuff3 |
peachpuff4 |
peru |
pink |
| pink1 |
pink2 |
pink3 |
pink4 |
| plum |
plum1 |
plum2 |
plum3 |
| plum4 |
powderblue |
purple |
purple1 |
| purple2 |
purple3 |
purple4 |
red |
| red1 |
red2 |
red3 |
red4 |
| rosybrown |
rosybrown1 |
rosybrown2 |
rosybrown3 |
| rosybrown4 |
royalblue |
royalblue1 |
royalblue2 |
| royalblue3 |
royalblue4 |
saddlebrown |
salmon |
| salmon1 |
salmon2 |
salmon3 |
salmon4 |
| sandybrown |
seagreen |
seagreen1 |
seagreen2 |
| seagreen3 |
seagreen4 |
seashell |
seashell1 |
| seashell2 |
seashell3 |
seashell4 |
sienna |
| sienna1 |
sienna2 |
sienna3 |
sienna4 |
| skyblue |
skyblue1 |
skyblue2 |
skyblue3 |
| skyblue4 |
slateblue |
slateblue1 |
slateblue2 |
| slateblue3 |
slateblue4 |
slategray |
slategray1 |
| slategray2 |
slategray3 |
slategray4 |
slategrey |
| snow |
snow1 |
snow2 |
snow3 |
| snow4 |
springgreen |
springgreen1 |
springgreen2 |
| springgreen3 |
springgreen4 |
steelblue |
steelblue1 |
| steelblue2 |
steelblue3 |
steelblue4 |
tan |
| tan1 |
tan2 |
tan3 |
tan4 |
| thistle |
thistle1 |
thistle2 |
thistle3 |
| thistle4 |
tomato |
tomato1 |
tomato2 |
| tomato3 |
tomato4 |
turquoise |
turquoise1 |
| turquoise2 |
turquoise3 |
turquoise4 |
violet |
| violetred |
violetred1 |
violetred2 |
violetred3 |
| violetred4 |
wheat |
wheat1 |
wheat2 |
| wheat3 |
wheat4 |
whitesmoke |
yellow |
| yellow1 |
yellow2 |
yellow3 |
yellow4 |
| yellowgreen |
|
Regular Expression
R has various functions for regular expression based match and replaces. The grep, grepl, regexpr and gregexpr functions are used for searching
for matches, while sub and gsub for performing replacement.
• grep(value = FALSE) returns an integer vector of the indices
of the elements of x that yielded a match (or not, for
invert = TRUE).
>str <- c("Regular", "expression", "examples of R language")
>x <- grep("ex",str,value=F)
>x
[1] 2 3
>x <- "line 4322: He is now 25 years old, and weights 130lbs";
>x <- grep("\\d","",x)
>x
[1] 1
• grep(value = TRUE) returns a character vector containing the
selected elements of x (after coercion, preserving names but no
other attributes).
>x <- grep("ex",str,value=T)
>x
[1] "expression" "examples of R language"
• grepl returns a logical vector (match or not for each element of
x).
>x <- grepl("ex",str)
>x
[1] FALSE TRUE TRUE
• sub and gsub return a character vector of the same
length and with the same attributes as x (after possible
coercion to character). Elements of character vectors x which
are not substituted will be returned unchanged (including any declared
encoding). If useBytes = FALSE a non-ASCII substituted result
will often be in UTF-8 with a marked encoding (e.g. if there is a
UTF-8 input, and in a multibyte locale unless fixed = TRUE).
>str <- c("Regular", "expression", "examples of R language")
>x <- sub("x.ress","",str)
>x
[1] "Regular" "eion" "examples of R language"
>x <- sub("x.+e","",str)
>x
[1] "Regular" "ession" "e"
>x <- "line 4322: He is now 25 years old, and weights 130lbs";
>x <- gsub("[[:digit:]]","",x)
>x
[1] "line : He is now years old, and weights lbs"
>x <- "line 4322: He is now 25 years old, and weights 130lbs";
>x <- gsub("\\d+","",x)
>x
[1] "line : He is now years old, and weights lbs"
• regexpr returns an integer vector of the same length as
text giving the starting position of the first match or
-1 if there is none, with attribute "match.length", an
integer vector giving the length of the matched text (or -1 for
no match). The match positions and lengths are in characters unless
useBytes = TRUE is used, when they are in bytes.
>str <- c("Regular", "expression", "examples of R language")
>x <- regexpr("x*ress",str)
>x
[1] -1 4 -1
• gregexpr returns a list of the same length as text each
element of which is of the same form as the return value for regexpr,
except that the starting positions of every (disjoint) match are
given.
>str <- c("Regular", "expression", "examples of R language")
>x <- gregexpr("x*ress",str)
>x
[[1]]
[1] -1
attr(,"match.length")
[1] -1
attr(,"useBytes")
[1] TRUE
[[2]]
[1] 4
attr(,"match.length")
[1] 4
attr(,"useBytes")
[1] TRUE
[[3]]
[1] -1
attr(,"match.length")
[1] -1
attr(,"useBytes")
[1] TRUE
Function Syntax:
grep(pattern, x, ignore.case = FALSE, perl = FALSE, value = FALSE,
fixed = FALSE, useBytes = FALSE, invert = FALSE)
grepl(pattern, x, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
sub(pattern, replacement, x, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
gsub(pattern, replacement, x, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
regexpr(pattern, text, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
gregexpr(pattern, text, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
Regular Expression Syntax:
| Syntax | Description |
| \\d | Digit, 0,1,2 ... 9 |
| \\D | Not Digit |
| \\s | Space |
| \\S | Not Space |
| \\w | Word |
| \\W | Not Word |
| \\t | Tab |
| \\n | New line |
| ^ | Beginning of the string |
| $ | End of the string |
| \ | Escape special characters, e.g. \\ is "\", \+ is "+" |
| | | Alternation match. e.g. /(e|d)n/ matches "en" and "dn" |
| • | Any character, except \n or line terminator |
| [ab] | a or b |
| [^ab] | Any character except a and b |
| [0-9] | All Digit |
| [A-Z] | All uppercase A to Z letters |
| [a-z] | All lowercase a to z letters |
| [A-z] | All Uppercase and lowercase a to z letters |
| i+ | i at least one time |
| i* | i zero or more times |
| i? | i zero or 1 time |
| i{n} | i occurs n times in sequence |
| i{n1,n2} | i occurs n1 - n2 times in sequence |
| i{n1,n2}? | non greedy match, see above example |
| i{n,} | i occures >= n times |
| [:alnum:] | Alphanumeric characters: [:alpha:] and [:digit:] |
| [:alpha:] | Alphabetic characters: [:lower:] and [:upper:] |
| [:blank:] | Blank characters: e.g. space, tab |
| [:cntrl:] | Control characters |
| [:digit:] | Digits: 0 1 2 3 4 5 6 7 8 9 |
| [:graph:] | Graphical characters: [:alnum:] and [:punct:] |
| [:lower:] | Lower-case letters in the current locale |
| [:print:] | Printable characters: [:alnum:], [:punct:] and space |
| [:punct:] | Punctuation character: ! " # $ % & ' ( ) * + , - . / : ; < = > ? @ [ \ ] ^ _ ` { | } ~ |
| [:space:] | Space characters: tab, newline, vertical tab, form feed, carriage return, space |
| [:upper:] | Upper-case letters in the current locale |
| [:xdigit:] | Hexadecimal digits: 0 1 2 3 4 5 6 7 8 9 A B C D E F a b c d e f |
Plot PCH Symbols Chart
Following is a chart of PCH symbols used in R plot. When the PCH is 21-25, the parameter "col=" and "bg=" should be specified.
PCH can also be in characters, such as "#", "%", "A", "a", and the character will be ploted.

Values pch=26:32 are currently unused, and pch=32:255 give the text symbol in a single-byte locale. In a multi-byte locale such as UTF-8, numeric values of pch greater than or equal to 32 specify a Unicode code point.
If pch is an integer or character NA or an empty character string, the point is omitted from the plot.
Value pch="." is handled specially. It is a rectangle of side 0.01 inch (scaled by cex). In addition, if cex = 1 (the default), each side is at least one pixel (1/72 inch on the pdf, postscript and xfig devices).
pch=0,square
pch=1,circle
pch=2,triangle point up
pch=3,plus
pch=4,cross
pch=5,diamond
pch=6,triangle point down
pch=7,square cross
pch=8,star
pch=9,diamond plus
pch=10,circle plus
pch=11,triangles up and down
pch=12,square plus
pch=13,circle cross
pch=14,square and triangle down
pch=15, filled square blue
pch=16, filled circle blue
pch=17, filled triangle point up blue
pch=18, filled diamond blue
pch=19,solid circle blue
pch=20,bullet (smaller circle)
pch=21, filled circle red
pch=22, filled square red
pch=23, filled diamond red
pch=24, filled triangle point up red
pch=25, filled triangle point down red
By default, pch=1 if not specified, and in black color:
>x <- c(2,1,3,2,5,3.3,1.4);
>y <- c(4,2.7,6,3,8,6,2.2);
>plot(x,y)

cex controls the symbol size in the plot, default is
cex=1,
col controls the color of the symbol border, default is
col="black".
Plot with specified PCH, Color and Size:
>plot(x,y,pch=2,cex=4,col="red")

Plot Function
plot(...) is a generic X Y plotting function. It's usage is:
plot(x, y = NULL, type = "p", xlim = NULL, ylim = NULL,
log = "", main = NULL, sub = NULL, xlab = NULL, ylab = NULL,
ann = par("ann"), axes = TRUE, frame.plot = axes,
panel.first = NULL, panel.last = NULL, asp = NA, ...)
x,y:Vector of coordinates
First let's make a simple plot:
>x <- c(1.2,3.4,1.3,-2.1,5.6,2.3,3.2,2.4,2.1,1.8,1.7,2.2)
>y <- c(2.4,5.7,2.0,-3,13,5,6.2,4.8,4.2,3.5,3.7,5.2)
>plot(x,y)

The
par(...) controls the general layout of the plot. For example,
par(mar = c(5, 4, 2, 1)) defines the bottom margin as 5, left margin 4,
top margin 2 and right margin as 1.
The default type is a point plot (type="p"). The possible types include:
| p: | Points, default |
| l: | Lines |
| b: | Points with line connection |
| c: | Line connections without points |
| o: | Both overplotted |
| h: | Histogram like vertical lines |
| s: | Stair steps |
| S: | Stair steps, another style |
| n: | No plotting |
Let's use less points and plot with line connections. We will
use blue colored line and points, and with axis labels both to X and Y axis as well as a main title of the plot:
>x <- c(-2,-0.3,1.4,2.4,4.5)
>y <- c(5,-0.5,8,2,11)
>par(mar = c(5, 1, 1, 1))
>plot(x,y,type="l",col="blue",xlab="Advertise Change",
+ ylab="Revenue Change", main="Financial Analysis")

Add more data to the plot:
>abline(v=0,col="red") #add a vertical line at x=0
>points(c(1,4),c(9,2),pch=3,col="tan2") #add two points
>x2 <- c(-1.5,1,4)
>y2 <- c(3,2,8)
>lines(x2,y2,col="darkolivegreen3") #add aother group of data
>legend(x=-2.2,y=11,c("advertise","sale"),cex=.8,
+ col=c("blue","darkolivegreen3"),lty=c(1,1)) #add legend

If we want to move the legend out of the main plot area, we need some more work.
First use
layout(...) function to define 2 plots on one layer side by side,
and then we plot the same data on both plots, with the plot on the right side in white color,
thus invisible (just providing the scale), and finally we plot the legend on the second plot.
>x <- c(-2,-0.3,1.4,2.4,4.5)
>y <- c(5,-0.5,8,2,11)
>layout(matrix(c(1,2), nrow = 1), widths = c(0.6, 0.4))
>par(mar = c(5, 4, 2, 1))
>plot(x,y,type="b",col="blue",xlab="Advertise Change",
+ ylab="Revenue Change", main="Financial Analysis")
>abline(v=0,col="red") #add a vertical line at x=0
>points(c(1,4),c(9,2),pch=3,col="tan2") #add two points
>x2 <- c(-1.5,1,4)
>y2 <- c(3,2,8)
>lines(x2,y2,col="darkolivegreen3")
>par(mar = c(5, 0, 2, 1))
>plot(x,y,col="white",axes=FALSE,ann=FALSE)
>legend(x=-2.2,y=11,c("advertise","sale"),cex=.8,
+ col=c("blue","darkolivegreen3"),lty=c(1,1))

String Functions
R string functions include substr(x), nchar(x), toupper(x), tolower(x),
strsplit(x,y),paste(...), and regular expression functions sub(...), grep(...) etc.
>s <- "EndMemo.com R Language Tutorial"
>substr(s,0,7)
[1] "EndMemo"
Get string length:
>nchar(s)
[1] 31
To uppercase:
>x <- toupper(s)
>x
[1] "ENDMEMO.COM R LANGUAGE TUTORIAL"
To lowercase:
>x <- tolower(s)
>x
[1] "endmemo.com r language tutorial"
Split the string at letter "o":
>x <- strsplit(s,"o")
[[1]]
[1] "EndMem" ".c" "m R Language Tut" "rial"
Concatenate two strings:
>x <- paste(x," -- String Functions",sep="")
>x
[1] "endmemo.com r language tutorial -- String Functions"
Substring replacement:
>x <- sub("Tutorial","Examples",s)
>x
[1] "EndMemo.com R Language Examples"
Use regular expression:
>x <- sub("n.+e","XXX",s)
>x
[1] "EXXX Tutorial"
Please see
grep() function for more regular expression handling of string.
tapply Function
tapply() applies a function to each cell of a ragged array.
tapply(X, INDEX, FUN = NULL, ..., simplify = TRUE)
•
X: vector
•
INDEX: list of one of more factors
•
FUN: the function
•
simplify: if true, return an array of scalar, other wise an array of list
...
>Orange #R built-in dataset, Growth of Orange Trees
Tree age circumference
1 1 118 30
2 1 484 58
3 1 664 87
4 1 1004 115
5 1 1231 120
6 1 1372 142
7 1 1582 145
8 2 118 33
9 2 484 69
10 2 664 111
11 2 1004 156
12 2 1231 172
13 2 1372 203
14 2 1582 203
15 3 118 30
16 3 484 51
17 3 664 75
18 3 1004 108
19 3 1231 115
20 3 1372 139
21 3 1582 140
22 4 118 32
23 4 484 62
24 4 664 112
25 4 1004 167
26 4 1231 179
27 4 1372 209
28 4 1582 214
29 5 118 30
30 5 484 49
31 5 664 81
32 5 1004 125
33 5 1231 142
34 5 1372 174
35 5 1582 177
Calculate the mean circumference of different Tree groups:
> tapply(Orange$circumference,Orange$Tree,mean)
3 1 5 2 4
94.00000 99.57143 111.14286 135.28571 139.28571
Return a list:
> tapply(Orange$circumference,Orange$Tree,mean,simplify=FALSE)
$`3`
[1] 94
$`1`
[1] 99.57143
$`5`
[1] 111.1429
$`2`
[1] 135.2857
$`4`
[1] 139.2857
pushBack Function
pushBack() function push back text lines onto a connection, and to enquire how many lines are currently pushed back.
pushBack(data, con, newLine = TRUE)
pushBackLength(con)
data: character vector
con: connection
newLine: logical. If true, a newline is appended to each string pushed back
> zz <- textConnection(LETTERS)
> readLines(zz, 2)
[1] "A" "B"
> pushBack("r",zz)
> pushBackLength(zz)
[1] 1
> readLines(zz, 1)
[1] "r"
> pushBackLength(zz)
[1] 0
> readLines(zz,1)
[1] "C"
> close(zz)
Quantile-Quantile Plot Example
Quantile-Quantile plot is a popular method to display data by plot the quantiles of the values against the corresponding quantiles of the normal (bell shapes).
The quantiles of the standard normal distribution is represented by a straight line. The normality of the data can be evaluated by observing
the extent in which the points appear on the line.
Following is a csv file example, we will draw a Quantile-Quantile plot of "Expression" values:
Let first read in the data from the file:
> x <- read.csv("histogram.csv",header=T,sep="\t")
> x <- t(x)
> ex <- as.numeric(x[2,1:ncol(x)])
Draw a Quantile-Quantile plot:
> qqnorm(ex)
> qqline(ex,col="red")

The above plot shows that most of the data points are on or near the straight line, suggests that the data is almost normally distributed.
For further test of the data normality, we can check the mean and median of the dataset.
> mean(ex)
[1] -0.3053381
> median(ex)
[1] -0.29
Mean is the average of the values, and the median is the second quartile, when the data is normally distributed around the mean, then the mean
and median should be equal. Since the mean and median above (-0.3053381 vs -0.29) are very close, so the data is seems quite symmetric.
We can write the plot into a file:
> png("histogram3.png",400,300)
> qqnorm(ex)
> qqline(ex,col="red")
> graphics.off()
qr Function
qr() function computes the QR decomposition of a matrix. It provides an interface to the techniques used in the LINPACK routine DQRDC or the LAPACK routines DGEQP3 and (for complex matrices) ZGEQP3.
qr(x, ...)
qr(x, tol = 1e-07 , LAPACK = FALSE, ...)
qr.coef(qr, y)
qr.qy(qr, y)
qr.qty(qr, y)
qr.resid(qr, y)
qr.fitted(qr, y, k = qr$rank)
qr.solve(a, b, tol = 1e-7)
solve(a, b, ...)
is.qr(x)
as.qr(x)
x: matrix
tol: the tolerance for detecting linear dependencies in the columns of x. Only used if LAPACK is false and x is real
qr: a QR decomposition of the type computed by qr
y,b: a vector or matrix of right-hand sides of equations
a: a QR decomposition or (qr.solve only) a rectangular matrix
k: effective rank
LAPACK: logical. For real x, if true use LAPACK otherwise use LINPACK
...
quit Function
quit() function terminate the current R session.
quit(save = "default", status = 0, runLast = TRUE)
q(save = "default", status = 0, runLast = TRUE)
save: a character string indicating whether the environment (workspace) should be saved, one of "no", "yes", "ask" or "default"
status: the (numerical) error status to be returned to the operating system, where relevant. Conventionally 0 indicates successful completion
runLast: should .Last() be executed?
...
> q()
This will shout down the R session without warning:
> q(save="no")
Random Number Generation
.Random.seed is an integer vector, containing the random number generator (RNG) state for random number generation in R. It can be saved and restored, but should not be altered by the user.
RNGkind is a more friendly interface to query or set the kind of RNG in use.
RNGversion can be used to set the random generators as they were in an earlier R version (for reproducibility).
set.seed is the recommended way to specify seeds.
.Random.seed <- c(rng.kind, n1, n2, ...)
RNGkind(kind = NULL, normal.kind = NULL)
RNGversion(vstr)
set.seed(seed, kind = NULL, normal.kind = NULL)
kind: character or NULL. If kind is a character string, set R's RNG to the kind desired. Use "default" to return to the R default
normal.kind: character string or NULL. If it is a character string, set the method of Normal generation. Use "default" to return to the R default. NULL makes no change
seed: a single value, interpreted as an integer
vstr: a character string containing a version number
rng.kind: integer code in 0:k for the above kind
n1,n2: integers
> runif(1)
[1] 0.7588484
> runif(1)
[1] 0.2751473
> require(stats)
> .Random.seed[1:6]
[1] 403 2 979405417 1358566968 1660710630 -1736144255
range Function
range() function get a vector of the minimum and maximum values.
range(..., na.rm = FALSE, finite = FALSE)
...: numeric vector
na.rm: whether NA should be removed, if not, NA will be returned
finite: whether non-finite elements should be omitted
>x <- c(1,2.3,2,3,4,8,12,43,-4,-1)
>r <- range(x)
>r
[1] -4 43
>diff(r)
[1] 47
Missing value affect the results:
>y<- c(x,NA)
>y
[1] 1.0 2.3 2.0 3.0 4.0 8.0 12.0 43.0 -4.0 -1.0 NA
>range(y)
[1] NA NA
After define na.rm=TRUE, result is meaningful:
>range(y,na.rm=TRUE)
[1] -4 43
> range(y,finite=TRUE)
[1] -4 43
rank Function
rank() function returns the sample ranks of the values in a vector. Ties (i.e., equal values) and missing values can be handled in several ways.
rank(x, na.last = TRUE,
ties.method = c("average", "first", "random", "max", "min"))
x: numeric, complex, character or logical vector
na.last: for controlling the treatment of NAs. If TRUE, missing values in the data are put last; if FALSE, they are put first; if NA, they are removed; if "keep" they are kept with rank NA
ties.method: a character string specifying how ties are treated, see ‘Details’; can be abbreviated
> x <- c(3,5,1,-4,NA,Inf,90,43)
> rank(x)
[1] 3 4 2 1 8 7 6 5
> rank(x, na.last=FALSE)
[1] 4 5 3 2 1 8 7 6
raw Function
raw() function creates or tests for objects of type "raw".
raw(length = 0)
as.raw(x)
is.raw(x)
length: disired length
x: object to be tested
> x <- raw(2)
> x
[1] 00 00
> x <- raw(10)
> x
[1] 00 00 00 00 00 00 00 00 00 00
> is.raw(x)
[1] TRUE
> x <- c(rep(1:8))
> x
[1] 1 2 3 4 5 6 7 8
> is.raw(x)
[1] FALSE
> as.raw(x)
[1] 01 02 03 04 05 06 07 08
> y <- as.raw(x)
> is.raw(y)
[1] TRUE
rawConnection Function
rawConnection() function inputs and outputs raw connections.
rawConnection(object, open = "r")
rawConnectionValue(con)
object: character or raw vector. A description of the connection. For an input this is an R raw vector object, and for an output connection the name for the connection
open: open mode
con: an output raw connection
> zz <- rawConnection(raw(0), "r+")
> writeBin(LETTERS,zz)
> seek(zz,0)
[1] 52
> readLines(zz)
[1] "A"
Warning message:
In readLines(zz) : incomplete final line found on 'raw(0)'
> seek(zz,0)
[1] 52
> writeBin(letters[1:3],zz)
> rawConnectionValue(zz)
[1] 61 00 62 00 63 00 44 00 45 00 46 00 47 00 48 00
49 00 4a 00 4b 00 4c 00 4d
[26] 00 4e 00 4f 00 50 00 51 00 52 00 53 00 54 00 55
00 56 00 57 00 58 00 59 00
[51] 5a 00
> close(zz)
rbind Function
rbind() function combines vector, matrix or data frame by rows.
rbind(x1,x2,...)
x1,x2:vector, matrix, data frames
data1.csv:
data2.csv:
Read in the data from the file:
>x <- read.csv("data1.csv",header=T,sep=",")
>x2 <- read.csv("data2.csv",header=T,sep=",")
>x3 <- rbind(x,x2)
>x3
Subtype Gender Expression
1 A m -0.54
2 A f -0.80
3 B f -1.03
4 C m -0.41
5 D m 3.22
6 D f 1.02
7 D f 0.21
8 D m -0.04
9 D m 2.11
10 B m -1.21
11 A f -0.20
The column of the two datasets must be same, otherwise the combination will be meaningless.
Read.csv Example
read.csv() function reads a file into data frame.
CSV file can be comma delimited or tab or any other delimiter specified by parameter "sep=".
If the parameter "header=" is "TRUE", then the first row will be treated as the row names.
read.csv(file, header = FALSE, sep = ",", quote = "\"",
dec = ".", fill = TRUE, comment.char = "", ...)
read.csv2(file, header = TRUE, sep = ";", quote = "\"",
dec = ",", fill = TRUE, comment.char = "", ...)
•
file: file name
•
header: 1st line as header or not, logical
•
sep: field separator
•
quote: quoting characters
...
The difference between read.csv and read.csv2 is the default field seperator, as "," and ";" respectively.
Following is a csv file example:
> x <- read.csv("readcsv.csv", header=T, dec=".",sep="\t")
> typeof(x)
[1] "list"
> is.data.frame(x)
[1] TRUE
We need to summarize how many 0, 1 and 2 are there in sample t1, t2 ... t8.
Following R code can be used to handle the job:
R source file:
x <- read.csv("readcsv.csv", header=T, dec=".",sep="\t")
xC = ncol(x)
xR = nrow(x)
ll <- data.frame(matrix(data = 0, nrow=3, ncol=8,byrow=T))
colnames(ll) <- names(x[,2:xC])
rownames(ll) <- c(0,1,2)
for (c in 2:xC)
{
for (r in 1:xR)
{
if (x[r,c]==0)
{
ll[1,c-1] = ll[1,c-1] + 1;
}
else if (x[r,c]==1)
{
ll[2,c-1] = ll[2,c-1] + 1;
}
else if (x[r,c]==2)
{
ll[3,c-1] = ll[3,c-1] + 1;
}
}
}
print(ll)
The result is:
t1 t2 t3 t4 t5 t6 t7 t8
0 9 10 11 5 5 7 10 8
1 7 3 6 11 10 8 6 8
2 4 7 3 4 5 5 4 4
Read.delim Example
read.delim() function reads a file into list.
The file by default is separated by tab, it can be comma delimited or any other delimiter specified by parameter "sep=".
If the parameter "header=" is "TRUE", then the first row will be treated as the row names.
read.delim(file, header = FALSE, sep = "\t", quote = "\"",
dec = ".", fill = TRUE, comment.char = "", ...)
read.delim2(file, header = TRUE, sep = "\t", quote = "\"",
dec = ",", fill = TRUE, comment.char = "", ...)
•
file: file name
•
header: 1st line as header or not, logical
•
sep: field separator
•
quote: quoting characters
...
read.delim() is almost the same as
read.table(), except the field separator is tab by default. It is convenient for
open tab delimited file.
Following is a csv file example:
> x <- read.delim("readcsv.csv", header=T)
> typeof(x)
[1] "list"
> is.data.frame(x)
[1] TRUE
Read.table Example
read.table() function reads a file into data frame in table format.
The file can be comma delimited or tab or any other delimiter specified by parameter "sep=".
If the parameter "header=" is "TRUE", then the first row will be treated as the row names.
read.table(file, header = FALSE, sep = "", quote = "\"'",
dec = ".", row.names, col.names,
as.is = !stringsAsFactors,
na.strings = "NA", colClasses = NA, nrows = -1,
skip = 0, check.names = TRUE, fill = !blank.lines.skip,
strip.white = FALSE, blank.lines.skip = TRUE,
comment.char = "#",
allowEscapes = FALSE, flush = FALSE,
stringsAsFactors = default.stringsAsFactors(),
fileEncoding = "", encoding = "unknown", text)
•
file: file name
•
header: 1st line as header or not, logical
•
sep: field separator
•
quote: quoting characters
...
Following is a csv file example "tp.txt":
> x <- read.table("tp.txt",header=T,sep="\t");
> is.data.frame(x)
[1] TRUE
> x
X t1 t2 t3 t4 t5 t6 t7 t8
1 r1 1 0 1 0 0 1 0 2
2 r2 1 2 2 1 2 1 2 1
3 r3 0 0 0 2 1 1 0 1
4 r4 0 0 1 1 2 0 0 0
5 r5 0 2 1 1 1 0 0 0
6 r6 2 2 0 1 1 1 0 0
7 r7 2 2 0 1 1 1 0 1
8 r8 0 2 1 0 1 1 2 0
9 r9 1 0 1 2 0 1 0 1
10 r10 1 0 2 1 2 2 1 0
11 r11 1 0 0 0 1 2 1 2
12 r12 1 2 0 0 0 1 2 1
13 r13 2 0 0 1 0 2 1 0
14 r14 0 2 0 2 1 2 0 2
15 r15 0 0 0 2 0 2 2 1
16 r16 0 0 0 1 2 0 1 0
17 r17 2 1 0 1 2 0 1 0
18 r18 1 1 0 0 1 0 1 2
19 r19 0 1 1 1 1 0 0 1
20 r20 0 0 2 1 1 0 0 1
> ncol(x)
[1] 9
> nrow(x)
[1] 20
> rownames(x)
[1] "1" "2" "3" "4" "5" "6" "7" "8" "9" "10" "11" "12" "13" "14" "15"
[16] "16" "17" "18" "19" "20"
regexpr Function
regexpr returns an integer vector of the same length as
text giving the starting position of the first match or
-1 if there is none, with attribute "match.length", an
integer vector giving the length of the matched text (or -1 for
no match). The match positions and lengths are in characters unless
useBytes = TRUE is used, when they are in bytes.
regexpr(pattern, text, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
•
pattern: regular expression, or string for fixed=TRUE
•
text: string, the character vector
•
ignore.case: case sensitive or not
•
perl: logical. Should perl-compatible regexps be used? Has priority over extended
•
fixed: logical. If TRUE, pattern is a string to be matched as is. Overrides all conflicting arguments
•
useBytes: logical. If TRUE the matching is done byte-by-byte rather than character-by-character
> x <- "line 4322: He is now 25 years old, and weights 130lbs"
> y <- regexpr("\\d+",x)
> y
[1] 6
attr(,"match.length")
[1] 4
attr(,"useBytes")
[1] TRUE
> x <- "line 4322: He is now 25 years old, and weights 130lbs"
> y <- regexpr("[[:digit:]]",x)
> y
[1] 6
attr(,"match.length")
[1] 1
attr(,"useBytes")
[1] TRUE
> if (y[[1]][1] != -1) print("match")
[1] "match"
Vector match:
>str <- c("Regular", "expression", "examples of R language")
>x <- regexpr("x*ress",str)
>x
[1] -1 4 -1
Regular Expression Syntax:
| Syntax | Description |
| \\d | Digit, 0,1,2 ... 9 |
| \\D | Not Digit |
| \\s | Space |
| \\S | Not Space |
| \\w | Word |
| \\W | Not Word |
| \\t | Tab |
| \\n | New line |
| ^ | Beginning of the string |
| $ | End of the string |
| \ | Escape special characters, e.g. \\ is "\", \+ is "+" |
| | | Alternation match. e.g. /(e|d)n/ matches "en" and "dn" |
| • | Any character, except \n or line terminator |
| [ab] | a or b |
| [^ab] | Any character except a and b |
| [0-9] | All Digit |
| [A-Z] | All uppercase A to Z letters |
| [a-z] | All lowercase a to z letters |
| [A-z] | All Uppercase and lowercase a to z letters |
| i+ | i at least one time |
| i* | i zero or more times |
| i? | i zero or 1 time |
| i{n} | i occurs n times in sequence |
| i{n1,n2} | i occurs n1 - n2 times in sequence |
| i{n1,n2}? | non greedy match, see above example |
| i{n,} | i occures >= n times |
| [:alnum:] | Alphanumeric characters: [:alpha:] and [:digit:] |
| [:alpha:] | Alphabetic characters: [:lower:] and [:upper:] |
| [:blank:] | Blank characters: e.g. space, tab |
| [:cntrl:] | Control characters |
| [:digit:] | Digits: 0 1 2 3 4 5 6 7 8 9 |
| [:graph:] | Graphical characters: [:alnum:] and [:punct:] |
| [:lower:] | Lower-case letters in the current locale |
| [:print:] | Printable characters: [:alnum:], [:punct:] and space |
| [:punct:] | Punctuation character: ! " # $ % & ' ( ) * + , - . / : ; < = > ? @ [ \ ] ^ _ ` { | } ~ |
| [:space:] | Space characters: tab, newline, vertical tab, form feed, carriage return, space |
| [:upper:] | Upper-case letters in the current locale |
| [:xdigit:] | Hexadecimal digits: 0 1 2 3 4 5 6 7 8 9 A B C D E F a b c d e f |
remove Function
remove() and rm() function delete R objects.
remove(..., list = character(), pos = -1,
envir = as.environment(pos), inherits = FALSE)
rm (..., list = character(), pos = -1,
envir = as.environment(pos), inherits = FALSE)
...: objects to be removed
list: character vector naming objects to be removed
pos: where to do the removal. By default, uses the current environment
envir: enviroment to use
inherits: should the enclosing frames of the environment be inspected?
> x <- 3
> x
[1] 3
> rm(x)
> x
Error: object 'x' not found
Delete all objects in current enviroment:
> ls()
[1] "y" "zz"
> rm(list=ls())
> ls()
character(0)
rep Function
rep() function replicates the values in x.
rep(x, ...)
rep.int(x, times)
x: numeric vector
...: arguments including times (default = 1), length.out, each (each elements how many times)
>x <- rep(1:5)
[1] 1 2 3 4 5
Repeat 1 -5 two times:
>x <- rep(1:5,2)
[1] 1 2 3 4 5 1 2 3 4 5
Convert to a 5 × 2 matrix:
>dim(x) <- c(5,2)
>x
[,1] [,2] [,3] [,4] [,5]
[1,] 1 3 5 2 4
[2,] 2 4 1 3 5
Each element replicates two times:
x <- rep(1:5,each=2)
[1] 1 1 2 2 3 3 4 4 5 5
Convert to a 5 × 2 matrix:
>dim(x) <- c(5,2)
>x
[,1] [,2] [,3] [,4] [,5]
[1,] 1 2 3 4 5
[2,] 1 2 3 4 5
> rep.int(1:5,2)
[1] 1 2 3 4 5 1 2 3 4 5
repeat
repeat is similar to while and for loop, it will execute a block of commands repeatly till
break.
> total <- 0
> repeat { total <- total + 1; print(total); if (total > 6) break; }
[1] 1
[1] 2
[1] 3
[1] 4
[1] 5
[1] 6
[1] 7
> total
[1] 7
replace Function
replace() function replaces the values in x with indices given in list by those given in values. If necessary, the values in values are recycled.
replace(x, list, values)
x: vector
list: indices
values: replacement values
> x <- c("green","red","yellow")
> x
[1] "green" "red" "yellow"
> y <- replace(x,1,"good")
> y
[1] "good" "red" "yellow"
> y <- replace(x,c(1,2),c("good","second"))
> y
[1] "good" "second" "yellow"
Reserved Words
The reserved words in R's parser includes:
if, else, repeat, while, function, for, in, next, break
TRUE, FALSE, NULL, Inf, NaN, NA, NA_integer, NA_real,
NA_complex, NA_character
Trace Copying of Objects
tracemem() function marks an object so that a message is printed whenever the internal function duplicate is called. This happens when two objects share the same memory and one of them is modified. It is a major cause of hard-to-predict memory use in R.
tracemem(x)
untracemem(x)
retracemem(x, previous = NULL)
x: R object
previous: value as returned by tracemem or retracemem
> x <- 3
> tracemem(x)
[1] "<0x000000000f479148"
> y <- x
> untracemem(x)
> y
[1] 3
rev Function
rev() function reverses an R object, including vector, array etc.
rev(x)
x: vector
> x <- c("green","red","yellow")
> x
[1] "green" "red" "yellow"
> y <- rev(x)
> y
[1] "yellow" "red" "green"
> x <- c(rep(1:10))
> x
[1] 1 2 3 4 5 6 7 8 9 10
> rev(x)
[1] 10 9 8 7 6 5 4 3 2 1
rle Function
rle() function computes the lengths and values of runs of equal values in a vector – or the reverse operation.
rle(x)
inverse.rle(x, ...) #inverse function of rle()
x: an atomic vector for rle(); an object of class "rle" for inverse.rle()
...
> x <- c(rep(1:10))
> x
[1] 1 2 3 4 5 6 7 8 9 10
> rle(x)
Run Length Encoding
lengths: int [1:10] 1 1 1 1 1 1 1 1 1 1
values : int [1:10] 1 2 3 4 5 6 7 8 9 10
row Function
row() function returns a matrix of integers indicating their row number in a matrix-like object, or a factor indicating the row labels.
row(x, as.factor=FALSE)
x: matrix
as.factor: whether the value should be returned as a factor of row labels (created if necessary) rather than as numbers
> x <- matrix(1:9,3,3)
> x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> y <- row(x)
> y
[,1] [,2] [,3]
[1,] 1 1 1
[2,] 2 2 2
[3,] 3 3 3
sample Function
sample() function takes a sample of the specified size from the elements of x using either with or without replacement.
sample(x, size, replace = FALSE, prob = NULL)
sample.int(n, size = n, replace = FALSE, prob = NULL)
x: either a vector of one or more elements from which to choose, or a positive integer
n: a positive number, the number of items to choose from
size: a non-negative integer giving the number of items to choose
replace: Should sampling be with replacement
prob: a vector of probability weights for obtaining the elements of the vector being sampled
> x <- 1:8
> sample(x)
[1] 8 4 7 2 3 6 5 1
> sample(x,replace=TRUE)
[1] 7 6 2 4 1 3 1 1
> sample(c(0,1),12,replace=TRUE)
[1] 1 1 1 0 1 0 0 0 1 1 0 0
Significance Analysis of Microarrays (samr)
Significance Analysis of Microarray (SAM) can be done by the 'samr' package. To install the package:
> source("http://bioconductor.org/biocLite.R")
> biocLite("samr")
Suppose we have a log2 transformed microarray file named "samr.csv"(can be downloaded at the end of the artical), which have 48 samples.
The first 24 samples have phenotype A, and the other 24 samples have phenotype B. We are going to find out the Differentially Expressed Genes (DEGs)
between these two groups.
Let first read in the data from the file:
> x1 = read.csv("samr.csv",header=T,sep=",",dec=".")
Precess the data for 'samr' use:
> xcol <- ncol(x1)
> xrow <- nrow(x1)-1
> x2 <- x1[2:xrow,2:xcol]
> y1 <- c(rep(1,24), rep(2,24))
> data=list(x=as.matrix(x2),y=y1,logged2=TRUE)
Calculate all significant genes:
> samr.obj<-samr(data,resp.type="Two class unpaired", nperms=100)
> delta.table <- samr.compute.delta.table(samr.obj, min.foldchange=0.1,nvals=200)
> siggenes.table <- samr.compute.siggenes.table(samr.obj, del=0, data, delta.table,all.genes=TRUE)
Note: the "min.foldchange=0.1" means that the fold change for the two groups should be >0.1.
Let's write all FDR < 10% DEGs into a file (the 8th column of siggenes.table is FDR) :
> a <- siggenes.table$genes.up; # all up regulated genes
> b <- siggenes.table$genes.lo; # all down regulated genes
> c <- rbind(a,b)
> lo <- c[as.numeric(c[,8])<10,]
> for (i in 1:nrow(lo))
> {
> tp <- as.numeric(as.vector(as.matrix(lo[i,1])))-1;
> lo[i,3] <- as.character(as.vector(as.matrix(x1[tp,1])));
> }
> write.csv(lo,"DEGs_samr.csv")
sapply Function
sapply() function applies a function to margins of an array or matrix.
sapply(x, func, ..., simplify = TRUE, USE.NAMES = TRUE)
•
x: array
•
func: the function
...
>BOD #R built-in dataset, Biochemical Oxygen Demand
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
Sum up for each row:
> sapply(BOD, sum)
Time demand
22 89
Multipy all values by 10:
> sapply(BOD,function(x) 10 * x)
Time demand
[1,] 10 83
[2,] 20 103
[3,] 30 190
[4,] 40 160
[5,] 50 156
[6,] 70 198
Used for array, margin set to 1:
> x <- array(1:9)
> sapply(x,function(x) x * 10)
[1] 10 20 30 40 50 60 70 80 90
Two dimension array, margin can be 1 or 2:
> x <- array(1:9,c(3,3))
> x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> sapply(x,function(x) x * 10)
[1] 10 20 30 40 50 60 70 80 90
save Function
save() function writes an external representation of R objects to the specified file. The objects can be read back from the file at a later date by using the function load (or data in some cases).
save(..., list = character(),
file = stop("'file' must be specified"),
ascii = FALSE, version = NULL, envir = parent.frame(),
compress = !ascii, compression_level,
eval.promises = TRUE, precheck = TRUE)
save.image(file = ".RData", version = NULL, ascii = FALSE,
compress = !ascii, safe = TRUE)
...: the names of the objects to be saved (as symbols or character strings)
list: a character vector containing the names of objects to be saved
file: a (writable binary-mode) connection or the name of the file where the data will be saved (when tilde expansion is done). Must be a file name for version = 1
ascii: if TRUE, an ASCII representation of the data is written. The default value of ascii is FALSE which leads to a binary file being written
version: the workspace format version to use. NULL specifies the current default format. The version used from R 0.99.0 to R 1.3.1 was version 1. The default format as from R 1.4.0 is version 2
envir: environment to search for objects to be saved
compress: logical or character string specifying whether saving to a named file is to use compression. TRUE corresponds to gzip compression, and (from R 2.10.0) character strings "gzip", "bzip2" or "xz" specify the type of compression. Ignored when file is a connection and for workspace format version 1
compression_level: integer: the level of compression to be used. Defaults to 6 for gzip compression and to 9 for bzip2 or xz compression
eval.promises: logical: should objects which are promises be forced before saving?
precheck: logical: should the existence of the objects be checked before starting to save (and in particular before opening the file/connection)? Does not apply to version 1 saves
safe: logical. If TRUE, a temporary file is used for creating the saved workspace. The temporary file is renamed to file if the save succeeds. This preserves an existing workspace file if the save fails, but at the cost of using extra disk space during the save
> x <- 3
> y <- list(a=TRUE,b="good")
> save(x,y,file="tp.RData")
> save.image()
> unlink("tp.RData")
saveRDS Function
saveRDS() function writes a single R object to a file, and to restore it.
readRDS() function reads the file.
saveRDS(object, file = "", ascii = FALSE, version = NULL,
compress = TRUE, refhook = NULL)
readRDS(file, refhook = NULL)
object: R object ot serialize
file: a connection or the name of the file where the R object is saved to or read from
ascii: a logical. If TRUE, an ASCII representation is written; otherwise (default except for text-mode connections), a binary one is used
version: the workspace format version to use. NULL specifies the current default version (2). Versions prior to 2 are not supported, so this will only be relevant when there are later versions
compress: a logical specifying whether saving to a named file is to use "gzip" compression, or one of "gzip", "bzip2" or "xz" to indicate the type of compression to be used. Ignored if file is a connection
refhook: a hook function for handling reference objects
> saveRDS(women, "women.rds")
> women2 <- readRDS("women.rds")
> identical(women, women2)
[1] TRUE
> con <- gzfile("women.rds")
> str(readRDS(con))
'data.frame': 15 obs. of 2 variables:
$ height: num 58 59 60 61 62 63 64 65 66 67 ...
$ weight: num 115 117 120 123 126 129 132 135 139 142 ...
close(con)
scale Function
scale() function centers and/or scales the columns of a numeric matrix.
scale(x, center = TRUE, scale = TRUE)
x: numeric matrix
center: either a logical value or a numeric vector of length equal to the number of columns of x
scale: either a logical value or a numeric vector of length equal to the number of columns of x
> x <- matrix(1:9,3,3)
> scale(x)
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> x
[,1] [,2] [,3]
[1,] -1 -1 -1
[2,] 0 0 0
[3,] 1 1 1
attr(,"scaled:center")
[1] 2 5 8
attr(,"scaled:scale")
[1] 1 1 1
scan Function
scan() function read data from screen or file.
scan(file = "", what = double(), nmax = -1, n = -1, sep = "",
quote = if(identical(sep, "\n")) "" else "'\"", dec = ".",
skip = 0, nlines = 0, na.strings = "NA",
flush = FALSE, fill = FALSE, strip.white = FALSE,
quiet = FALSE, blank.lines.skip = TRUE, multi.line = TRUE,
comment.char = "", allowEscapes = FALSE,
fileEncoding = "", encoding = "unknown", text
•
file: the name of a file, if "", then read in from stdin
•
what: type of data, including logical, integer, numeric, complex, character, raw
...
Following is a csv file example.
> x <- scan("ordermatrix.csv",what="character",skip=1,quiet=TRUE);
> x
[1] "r1,1,0,1,0,0,1,0,2" "r2,1,2,5,1,2,1,2,1" "r3,0,0,9,2,1,1,0,1"
[4] "r4,0,0,2,1,2,0,0,0" "r5,0,2,15,1,1,0,0,0" "r6,2,2,3,1,1,1,0,0"
[7] "r7,2,2,3,1,1,1,0,1"
> x <- scan("ordermatrix.csv",what="character",quiet=TRUE);
> x
[1] ",t1,t2,t3,t4,t5,t6,t7,t8" "r1,1,0,1,0,0,1,0,2"
[3] "r2,1,2,5,1,2,1,2,1" "r3,0,0,9,2,1,1,0,1"
[5] "r4,0,0,2,1,2,0,0,0" "r5,0,2,15,1,1,0,0,0"
[7] "r6,2,2,3,1,1,1,0,0" "r7,2,2,3,1,1,1,0,1"
> x <- scan("ordermatrix.csv",skip=1,nlines=1);
Read 1 item
> x
[1] "r1,1,0,1,0,0,1,0,2"
Read into a list:
> x <- scan("ordermatrix.csv",skip=1,
+ what = list("","","","","","","","",""))
[[1]]
[1] "r1,1,0,1,0,0,1,0,2"
[[2]]
[1] "r2,1,2,5,1,2,1,2,1"
[[3]]
[1] "r3,0,0,9,2,1,1,0,1"
[[4]]
[1] "r4,0,0,2,1,2,0,0,0"
[[5]]
[1] "r5,0,2,15,1,1,0,0,0"
[[6]]
[1] "r6,2,2,3,1,1,1,0,0"
[[7]]
[1] "r7,2,2,3,1,1,1,0,1"
[[8]]
[1] ""
[[9]]
[1] ""
Read data from screen if let the file name "", or just without any parameter:
> x <- scan("",what="int")
1: 43 #input 43 from the screen
2:
Read 1 item
> x
[1] "43"
> x <- scan("",what="int")
1: 43 #input 43 from the screen
2: 22
3: 67
4:
Read 3 items
> x
[1] "43" "22" "67"
Large data can be scanned in by just copy and paste, for example paste from EXCEL.
> x <- scan()
Then use "ctrl+v" to paste the data, the data type will be automatically determined.
Scatter Plots Example
( Scatter Plot Online )
Following is a csv file example, we will draw a Scatter Plot of the "Expression" and "Quality" values:
Let first read in the data from the file:
> x <- read.csv("scatterplot.csv",header=T,sep="\t")
> x <- t(x)
> ex <- as.numeric(x[2,1:ncol(x)])
> qu <- as.numeric(x[3,1:ncol(x)])
Draw a Scatter Plot:
> plot(ex,qu)

If we want to draw different subtype in different color and symbol, we need more work like follows:
> plot(ex,qu,col="white",xlab="Expression", ylab="Quality")
> points(ex[1:143],qu[1:143],col="red",pch=3,cex=.6) #Subtype A
> points(ex[144:218],qu[144:218],col="blue",pch=19,cex=.6) #Subtype B
> points(ex[219:ncol(x)],qu[219:ncol(x)],col="black",,pch=1,cex=.6) #Subtype C
> abline(lm(ex[144:218] ~ qu[144:218]),col="blue") #regression expression ~ quality of B

Following code can add a legend on the right:
> layout(matrix(c(1,2), nrow = 1), widths = c(0.7, 0.3))
> par(mar = c(5, 4, 4, 2) + 0.1)
> plot(ex,qu,col="white",xlab="Expression", ylab="Quality")
> points(ex[219:ncol(x)],qu[219:ncol(x)],col="black",,pch=1,cex=.6)
> points(ex[144:218],qu[144:218],col="blue",pch=19,cex=.6)
> points(ex[1:143],qu[1:143],col="red",cex=.6,pch=3)
> abline(lm(ex[144:218] ~ qu[144:218]),col="blue")
> par(mar = c(5, 0, 4, 1) + 0.1)
> plot(ex,qu,axes=FALSE,ann=FALSE, col="white")
> legend(x=-2.5,y=1.2,c("A (n=146)","B (n=77)","C (n=85)"),cex=.8, pch=c(1,19,3),col=c("black","blue", "red"))

R package "scatterplot3d" can be used to draw 3D scatter plots, to install this package:
> install.packages("scatterplot3d",repos="http://R-Forge.R-project.org")
To draw a 3D scatter plot based on the "Expression", "Quality" and "Height" values:
> library(scatterplot3d)
> hi <- as.numeric(x[4,1:ncol(x)])
> scatterplot3d(ex,qu,hi,pch=20,highlight.3d=T)

We can add more parameters like:
> scatterplot3d(ex,qu,hi,pch=20,highlight.3d=T,type="h")

Download the csv file and the R source code:
Data File
R Source Code File
SD SE Calculations
sd() function calculates the standard deviation.
sd(x, na.rm=FALSE)
x: numeric vector
na.rm: missing values should be removed or not
> x <- c(1,2.3,2,3,4,8,12,43,-4,-1)
> r <- sd(x)
> r
[1] 13.39602
The standard error equals sd/√
n:
> x <- c(1,2.3,2,3,4,8,12,43,-4,-1)
> se <- sd(x)/sqrt(length(x))
> se
[1] 4.236195
Calculate the SD of data frame (matrix):
>BOD #R Biochemical Oxygen Demand database
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
> apply(BOD,2,sd)
Time demand
2.160247 4.630623
search Function
search() function gets the list of attached packages in the R Search Path.
The default packages in the search path:
>search()
[1] ".GlobalEnv" "package:stats" "package:graphics"
[4] "package:grDevices" "package:utils" "package:datasets"
[7] "package:methods" "Autoloads" "package:base"
Attach BOD to the search path:
>attach(BOD)
>search()
[1] ".GlobalEnv" "BOD" "package:stats"
[4] "package:graphics" "package:grDevices" "package:utils"
[7] "package:datasets" "package:methods" "Autoloads"
[10] "package:base"
To list the full path of the packages:
>searchpath()
[1] ".GlobalEnv"
[2] "BOD"
[3] "C:/Program Files/R/R-2.15.2/library/stats"
[4] "C:/Program Files/R/R-2.15.2/library/graphics"
[5] "C:/Program Files/R/R-2.15.2/library/grDevices"
[6] "C:/Program Files/R/R-2.15.2/library/utils"
[7] "C:/Program Files/R/R-2.15.2/library/datasets"
[8] "C:/Program Files/R/R-2.15.2/library/methods"
[9] "Autoloads"
[10] "C:/PROGRA~1/R/R-215~1.2/library/base"
seek Function
seek(con, ...)
seek(con, where = NA, origin = "start", rw = "", ...)
isSeekable(con)
truncate(con, ...)
con: connection
where: file position, numeric
rw: read or write
origin: start, current or end
...
seq Function
seq() function generates a sequence of numbers.
seq(from = 1, to = 1, by = ((to - from)/(length.out - 1)),
length.out = NULL, along.with = NULL, ...)
•
from, to: begin and end number of the sequence
•
by: step, increment (Default is 1)
•
length.out: length of the sequence
...
Generate a sequence from -6 to 7:
> x <- seq(-6,7)
> x
[1] -6 -5 -4 -3 -2 -1 0 1 2 3 4 5 6 7
From -6 till 7, step=2:
> x <- seq(-6,7,by=2)
> x
[1] -6 -4 -2 0 2 4 6
Let's try smaller step:
> x <- seq(-2,2,by=0.3)
> x
[1] -2.0 -1.7 -1.4 -1.1 -0.8 -0.5 -0.2 0.1 0.4
0.7 1.0 1.3 1.6 1.9
Suppose we do not know the step, but we want 10 evenly distributed numbers from
-2 to 2:
> seq(-2,2,length.out=10)
[1] -2.0000000 -1.5555556 -1.1111111 -0.6666667 -0.2222222 0.2222222
[7] 0.6666667 1.1111111 1.5555556 2.0000000
Generate a sequence from 1 to 10, quick version:
> x <- seq(10)
> x
[1] 1 2 3 4 5 6 7 8 9 10
The generated sequence is a vector:
> is.vector(x)
[1] TRUE
> exp(x)
[1] 2.718282 7.389056 20.085537 54.598150 148.413159
[6] 403.428793 1096.633158 2980.957987 8103.083928 22026.465795
sequence Function
sequence(x) function creates a vector of length x with elements 1,2,3 ... x.
sequence(x)
x: number or numeric vector
> sequence(3)
[1] 1 2 3
> sequence(8)
[1] 1 2 3 4 5 6 7 8
> sequence(c(3,8))
[1] 1 2 3 1 2 3 4 5 6 7 8
serialize Function
serialize() function is a simple low-level interface for serializing to connections.
serialize(object, connection, ascii, version = NULL, refhook = NULL)
unserialize(connection, refhook = NULL)
object: R object to serialize
connection: an open connection or (for serialize) NULL or (for unserialize) a raw vector
ascii: a logical. If TRUE, an ASCII representation is written; otherwise binary one. The default is TRUE for a text-mode connection and FALSE otherwise
version: the workspace format version to use. NULL specifies the current default version (2). Versions prior to 2 are not supported, so this will only be relevant when there are later versions
refhook: a hook function for handling reference objects
> x <- serialize(BOD,NULL)
> x
[1] 58 0a 00 00 00 02 00 03 00 01 00 02 03 00 00 00 03 13 00 00 00 02
00 00 00
[26] 0e 00 00 00 06 3f f0 00 00 00 00 00 00 40 00 00 00 00 00 00 00 40
08 00 00
[51] 00 00 00 00 40 10 00 00 00 00 00 00 40 14 00 00 00 00 00 00 40 1c
00 00 00
[76] 00 00 00 00 00 00 0e 00 00 00 06 40 20 99 99 99 99 99 9a 40 24 99
99 99 99
[101] 99 9a 40 33 00 00 00 00 00 00 40 30 00 00 00 00 00 00 40 2f 33 33
33 33 33
[126] 33 40 33 cc cc cc cc cc cd 00 00 04 02 00 00 00 01 00 04 00 09 00
00 00 05
[151] 6e 61 6d 65 73 00 00 00 10 00 00 00 02 00 04 00 09 00 00 00 04 54
69 6d 65
[176] 00 04 00 09 00 00 00 06 64 65 6d 61 6e 64 00 00 04 02 00 00 00 01
00 04 00
[201] 09 00 00 00 09 72 6f 77 2e 6e 61 6d 65 73 00 00 00 0d 00 00 00 02
80 00 00
[226] 00 ff ff ff fa 00 00 04 02 00 00 00 01 00 04 00 09 00 00 00 05 63
6c 61 73
[251] 73 00 00 00 10 00 00 00 01 00 04 00 09 00 00 00 0a 64 61 74 61 2e
66 72 61
[276] 6d 65 00 00 04 02 00 00 00 01 00 04 00 09 00 00 00 09 72 65 66 65
72 65 6e
[301] 63 65 00 00 00 10 00 00 00 01 00 04 00 09 00 00 00 0c 41 31 2e 34
2c 20 70
[326] 2e 20 32 37 30 00 00 00 fe
> BOD
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
sign Function
sign() function returns a vector with the signs of the corresponding elements of x (the sign of a real number is 1, 0, or -1 if the number is positive, zero, or negative, respectively).
> x <- c(3,2,0,-19,32,-5)
> sign(x)
[1] 1 1 0 -1 1 -1
> sign(4)
[1] 1
> sign(-3)
[1] -1
sink Function
sink() function diverts R output to a connection.
sink(file = NULL, append = FALSE, type = c("output", "message"),
split = FALSE)
sink.number(type = c("output", "message"))
file: a writable connection or a character string naming the file to write to, or NULL to stop sink-ing
append: logical. If TRUE, output will be appended to file; otherwise, it will overwrite the contents of file
type: character. Either the output stream or the messages stream
split: logical: if TRUE, output will be sent to the new sink and to the current output stream, like the Unix program tee
sink.number() gets how many diversions are in use.
sink.number(type="message") gets the number of connection currently being used for error
messages.
> sink("tp.txt") #writ all output to file tp.txt
> for (i in 1:5) print(i);
> sink() #stop sinking, =sink(NULL)
In the tp.txt, the content is:
[1] 1
[1] 2
[1] 3
[1] 4
[1] 5
> sink.number()
[1] 0
> for (i in 1:5) print(i) #no sinking, then print to screen
[1] 1
[1] 2
[1] 3
[1] 4
[1] 5
> unlink("tp.txt") #delete the file tp.txt
solve Function
solve() function solves equation a %*% x = b for x, where b is a vector or matrix.
solve(a, b, tol, LINPACK = FALSE, ...)
•
a: coefficients of the equation
•
b: vector or matrix of the equation right side
•
tol: the tolerance for detecting linear dependencies in the columns of a
•
LINPACK: logical. Defunct and ignored
...
5x = 10, what's x?
>solve(5,10)
[1] 2
Let's see two variables examples:
3x + 2y = 8
x + y =2
What's x and y?
In above equations, matrix a is:
3 2
1 1
Matrix b is:
8
2
> a <- matrix(c(3,1,2,1),nrow=2,ncol=2)
> a
[,1] [,2]
[1,] 3 2
[2,] 1 1
> b <- matrix(c(8,2),nrow=2,ncol=1)
> b
[,1]
[1,] 8
[2,] 2
> solve(a,b)
[,1]
[1,] 4
[2,] -2
So x = 4, y = -2.
If b is absent, the default is a unit matrix.
> x <- stats::rnorm(16)
> dim(x) <- c(4,4)
> x
[,1] [,2] [,3] [,4]
[1,] -0.3017359 -0.4687800 0.66832626 0.003768864
[2,] -0.8327101 0.7754996 -0.04494932 1.900833149
[3,] -0.1948664 -0.9313664 -0.47685005 -0.123290962
[4,] 1.2502012 -1.0014304 1.61952675 1.119330272
> solve(x)
[,1] [,2] [,3] [,4]
[1,] -1.0175034 -0.23116550 -0.09488446 0.38553721
[2,] -0.2013479 0.03601077 -0.78443594 -0.14687844
[3,] 0.8975934 -0.08140970 -0.59455159 0.06973859
[4,] -0.3423730 0.40820022 0.26440712 0.23046715
Get the inverse matrix of matrix x:
> solve(x) %*% x
[,1] [,2] [,3] [,4]
[1,] 1.000000e+00 0.000000e+00 -2.220446e-16 2.775558e-16
[2,] 8.881784e-16 1.000000e+00 -8.881784e-16 2.220446e-16
[3,] -8.881784e-16 0.000000e+00 1.000000e+00 -4.440892e-16
[4,] 0.000000e+00 -2.775558e-17 2.775558e-17 1.000000e+00
sort Function
sort() function sorts a vector.
sort(x, decreasing = FALSE, na.last = NA, ...)
x: vector
decreasing: decrease or not
na.last: if TRUE, NAs are put at last position, FALSE at first, if NA, remove them (default)
...
Sort Vectors:
>x <- c(1,2.3,2,3,4,8,12,43,-4,-1,NA)
>sort(x)
[1] -4.0 -1.0 1.0 2.0 2.3 3.0 4.0 8.0 12.0 43.0
>sort(x,decreasing=TRUE)
[1] 43.0 12.0 8.0 4.0 3.0 2.3 2.0 1.0 -1.0 -4.0
>sort(x,decreasing=TRUE, na.last=TRUE)
[1] 43.0 12.0 8.0 4.0 3.0 2.3 2.0 1.0 -1.0 -4.0 NA
>sort(x,decreasing=TRUE, na.last=FALSE)
[1] NA 43.0 12.0 8.0 4.0 3.0 2.3 2.0 1.0 -1.0 -4.0
Matrix
R matrix is a two dimensional array. R has a lot of operator and functions that make matrix handling very convenient.
Matrix assignment:
>A <- matrix(c(3,5,7,1,9,4),nrow=3,ncol=2,byrow=TRUE)
>A
[,1] [,2]
[1,] 3 5
[2,] 7 1
[3,] 9 4
Matrix row and column count:
>rA <- nrow(A)
>rA
[1] 3
>cA <- ncol(A)
>cA
[1] 2
t(A) function returns a transposed matrix of A:
>B <- t(A)
>B
[,1] [,2] [,3]
[1,] 3 7 9
[2,] 5 1 4
Matrix multplication:
C <- A * A
C
[,1] [,2]
[1,] 9 25
[2,] 49 1
[3,] 81 16
Matrix Addition:
>C <- A + A
>C
[,1] [,2]
[1,] 6 10
[2,] 14 2
[3,] 18 8
Matrix subtraction (-) and division (/) operations ... ...
Sometimes a matrix need to be sorted by a specific column, which can be done by using
order() function.
Following is a csv file example.
Following R code will read in the above file into a matrix, and sort it by column 4, then write to a output file.
The result is:
Download the csv file and the R source code:
Data File
R Source Code File
Order() returns a permutation which rearranges its first argument
into ascending or descending order, breaking ties by further
arguments.
Usage:
order(..., na.last = TRUE, decreasing = FALSE)
Arguments:
...: a sequence of numeric, complex, character or logical vectors,
all of the same length, or a classed R object.
decreasing: logical. Should the sort order be increasing or
decreasing?
na.last: for controlling the treatment of 'NA's. If 'TRUE', missing
values in the data are put last; if 'FALSE', they are put
first; if 'NA', they are removed.
split Function
split() function divides the data in a vector. unsplit() funtion do the reverse.
split(x, f, drop = FALSE, ...)
split(x, f, drop = FALSE, ...) <- value
unsplit(value, f, drop = FALSE)
x: vector, data frame
f: indices
drop: discard non existing levels or not
...
Following file has been used for ANOVA analysis:
(
Download the data file)
Let first read in the data from the file:
>x <- read.csv("anova.csv",header=T,sep=",")
Split the "Expression" values into two groups based on "Gender" variable, "f" for female group,
and "m" for male group:
>g <- split(x$Expression, x$Gender)
>g
$f
[1] -0.66 -1.15 -0.30 -0.40 -0.24 -0.92 0.48 -1.68 -0.80 -0.55 -0.11 -1.26
[13] -0.11 0.13 0.81 0.45 0.74 -0.31 -0.18 -0.08 0.54 -0.35 0.38 -0.39
[25] -1.49 -0.77 -0.92 -0.35 0.26 -0.78 1.20 0.06 -0.68 -0.44 0.93 -0.35
[37] 0.11 -0.12 -0.22 0.29 -0.67 -0.03 -0.57 0.19 -1.80 -0.81 1.80 -0.99
[49] -2.22 -1.06 0.06 -1.68 -0.64 0.29 -0.13 -0.84 0.44 -1.32 -0.54 -0.05
[61] 0.23 0.38 0.35 0.30 -0.33 0.79 -0.06 -0.88 0.32 -0.45 0.21 -2.03
[73] 0.59 -0.92 -0.07 -0.39 -0.98 -0.11 -0.73 -1.01 -0.50 -0.16 -0.59 1.13
[85] 1.01 0.21 -0.21 -1.05 0.10 -1.81 -1.18 0.49 -1.74 -1.57 0.46 1.31
[97] 0.44 -2.08 -1.62 -1.53 0.03 -0.42 -1.86 -1.99 -0.25 -2.11 -0.93 0.42
[109] -1.13 -0.92 0.38 -2.01 1.42 0.10 -2.17 0.13 -1.75 -1.18 0.85 0.64
[121] 0.97 -0.72 -0.04 0.38 -1.87 -2.09 -1.54 0.09 -0.25 0.51 0.33 -1.29
[133] -0.51 -0.50 -0.52
$m
[1] -0.54 -0.80 -1.03 -0.41 -1.31 -0.43 1.01 0.14 1.42 -0.16 0.15 -0.62
[13] -0.42 -0.35 -0.42 0.32 -0.57 -0.07 -0.06 0.02 -0.39 -0.74 -0.09 -0.03
[25] 0.18 0.25 -0.39 -0.24 -0.30 0.25 -0.42 0.54 0.03 -0.66 0.30 -0.38
[37] -0.03 -0.62 0.14 -0.77 -0.09 -0.80 -0.41 -0.88 -0.27 -0.07 -1.60 -0.79
[49] -0.33 1.31 -0.33 -0.43 -0.92 -0.29 -1.02 0.41 -0.81 0.61 -0.63 -0.49
[61] 0.18 0.17 0.24 -0.12 -0.24 -0.26 1.48 0.04 -0.56 -1.12 -0.19 0.27
[73] -1.28 -0.38 -0.83 0.25 -0.14 0.29 0.18 0.44 -0.28 0.08 -0.29 -0.62
[85] -0.87 0.19 0.34 0.54 0.02 -0.39 1.25 -0.51 0.05 -0.36 -0.19 -0.10
[97] 0.08 -1.16 1.58 0.59 -0.19 0.56 -0.22 -0.77 -0.12 -0.76 0.35 -0.69
[109] -0.20 -0.44 -1.98 0.00 -0.54 -0.61 -1.39 0.44 0.20 -0.78 -0.96 -0.10
[121] 0.39 -1.11 -1.78 -1.46 1.00 -1.34 -0.72 -0.47 0.15 1.67 0.81 0.16
[133] -0.39 -0.40 1.18 -0.30 -1.91 -1.14 0.13 -0.34 -0.44 0.52 1.11 -0.89
[145] -0.17 -1.62
Calculate the length, mean value of each group:
>sapply(g,length)
f m
135 146
>sapply(g,mean)
f m
-0.3946667 -0.2227397
You may use lapply, return is a list:
>lapply(g,mean)
$f
[1] -0.3946667
$m
[1] -0.2227397
unsplit() function combines the groups:
>unsplit(g,x$Gender)
sqrt Function
sqrt() function computes the square root of a numeric vector.
sqrt(x)
x: numeric or complex vector, array
> sqrt(9)
[1] 3
> sqrt(-1)
[1] NaN
Warning message:
In sqrt(-1) : NaNs produced
> sqrt(3+5i)
[1] 2.101303+1.189738i
> sqrt(c(4,9,16))
[1] 2 3 4
strsplit Function
strsplit() function splits the elements of a character vector x into substrings according to the matches to substring split within them.
strsplit(x, split, fixed = FALSE, perl = FALSE, useBytes = FALSE)
x: character vector
split: character vector, separator
fixed: logical. If TRUE match split exactly, otherwise use regular expressions. Has priority over perl
perl: logical. Should perl-compatible regexps be used?
useBytes: logical. If TRUE the matching is done byte-by-byte rather than character-by-character, and inputs with marked encodings are not converted. This is forced (with a warning) if any input is found which is marked as "bytes"
> x <- "r tutorial"
> strsplit(x,NULL)
[[1]]
[1] "r" " " "t" "u" "t" "o" "r" "i" "a" "l"
> y <- strsplit(x,"t")
> y
[[1]]
[1] "r " "u" "orial"
> unlist(y)
[1] "r " "u" "orial"
strtoi Function
strtoi() function converts string to integers.
strtoi(x, base=0L)
x: character vector
base: integer between 2 and 36 inclusive, default is 0
For the default base = 0L, the base chosen from the string representation of that element of x, so different elements can have different bases (see the first example). The standard C rules for choosing the base are that octal constants (prefix 0 not followed by x or X) and hexadecimal constants (prefix 0x or 0X) are interpreted as base 8 and 16; all other strings are interpreted as base 10.
For a base greater than 10, letters a to z (or A to Z) are used to represent 10 to 35.
strtrim Function
strtrim() function trims character strings to specified display widths.
strtrim(x,width)
x: character vector
width: positive integer
> x <- "r tutorial"
> strtrim(x,3)
[1] "r t"
> x <- c("green","red","blue")
> y <- strtrim(x,1)
> y
[1] "g" "r" "b"
> y <- strtrim(x,c(1,2,3))
> y
[1] "g" "re" "blu"
structure Function
structure() function gets the attributes of an R object.
structure(.Data, ...)
.Data: object
...: attributes, specified in tag=value form, which will be attached to data
> x <- c("green","red","blue")
> structure(x)
[1] "green" "red" "blue"
> structure(BOD)
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
strwrap Function
strwrap() function wrap character strings to format paragraphs.
Each character string in the input is first split into paragraphs (or lines containing whitespace only). The paragraphs are then formatted by breaking lines at word boundaries. The target columns for wrapping lines and the indentation of the first and all subsequent lines of a paragraph can be controlled independently.
strwrap(x, width = 0.9 * getOption("width"), indent = 0,
exdent = 0, prefix = "", simplify = TRUE, initial = prefix)
x: character vector
width: a positive integer giving the target column for wrapping lines in the output
indent: a non-negative integer giving the indentation of the first line in a paragraph
exdent: a non-negative integer specifying the indentation of subsequent lines in paragraphs
prefix, initial: a character string to be used as prefix for each line except the first, for which initial is used
simplify: a logical. If TRUE, the result is a single character vector of line text; otherwise, it is a list of the same length as x the elements of which are character vectors of line text obtained from the corresponding element of x. (Hence, the result in the former case is obtained by unlisting that of the latter.)
> x <- paste(readLines(file.path(R.home("doc"), "COPYING")),
+ collapse = "\n")
> y <- unlist(strsplit(x,"\n"))
> z <- y[-(1:330)]
> z
[1] " `Gnomovision' (which makes passes at compilers) written
by James Hacker."
[2] ""
[3] " , 1 April 1989"
[4] " Ty Coon, President of Vice"
[5] ""
[6] "This General Public License does not permit incorporating
your program into"
[7] "proprietary programs. If your program is a subroutine
library, you may"
[8] "consider it more useful to permit linking proprietary
applications with the"
[9] "library. If this is what you want to do, use the GNU
Library General"
[10] "Public License instead of this License."
> strwrap(z, width=20)
[1] "`Gnomovision'" "(which makes passes" "at compilers)"
[4] "written by James" "Hacker." ""
[7] ", 1 April 1989" "Ty Coon, President"
[10] "of Vice" "" "This General Public"
[13] "License does not" "permit" "incorporating your"
[16] "program into" "proprietary" "programs. If your"
[19] "program is a" "subroutine library," "you may"
[22] "consider it more" "useful to permit" "linking proprietary"
[25] "applications with" "the" "library. If this"
[28] "is what you want to" "do, use the GNU" "Library General"
[31] "Public License" "instead of this" "License."
sub Function
sub() function replaces the first match of a string, if the parameter is a string vector, replaces
the first match of all elements.
sub(pattern, replacement, x, ignore.case = FALSE, perl = FALSE,
fixed = FALSE, useBytes = FALSE)
•
pattern: regular expression, or string for fixed=TRUE
•
x: string, the character vector
•
replacement: string, character vector for replacement
•
ignore.case: case sensitive or not
•
perl: logical. Should perl-compatible regexps be used? Has priority over extended
•
fixed: logical. If TRUE, pattern is a string to be matched as is. Overrides all conflicting arguments
•
useBytes: logical. If TRUE the matching is done byte-by-byte rather than character-by-character
> x <- "r tutorial"
> y <- sub("r ","HTML ", x)
> y
[1] "HTML tutorial"
> y <- sub("t.*r","BBBBB", x) #regular expression substitution
> y
[1] "r BBBBBial"
> y <- sub("t.*r","BBBBB", x, fixed=TRUE) #not regular expression
> y
[1] "r tutorial"
sub can be used for vector replacement. Following
example replaces one digit of all elements in the vector:
> x <- c("line 435", "good weather", "89 pigs")
> y <- sub("[[:digit:]]","",x)
> y
[1] "line 35" "good weather" "9 pigs"
Replace all digits of the vector elements:
> x <- c("line 435", "good weather", "89 pigs")
> y <- sub("[[:digit:]]+","",x)
> y
[1] "line " "good weather" " pigs"
Regular Expression Syntax:
| Syntax | Description |
| \\d | Digit, 0,1,2 ... 9 |
| \\D | Not Digit |
| \\s | Space |
| \\S | Not Space |
| \\w | Word |
| \\W | Not Word |
| \\t | Tab |
| \\n | New line |
| ^ | Beginning of the string |
| $ | End of the string |
| \ | Escape special characters, e.g. \\ is "\", \+ is "+" |
| | | Alternation match. e.g. /(e|d)n/ matches "en" and "dn" |
| • | Any character, except \n or line terminator |
| [ab] | a or b |
| [^ab] | Any character except a and b |
| [0-9] | All Digit |
| [A-Z] | All uppercase A to Z letters |
| [a-z] | All lowercase a to z letters |
| [A-z] | All Uppercase and lowercase a to z letters |
| i+ | i at least one time |
| i* | i zero or more times |
| i? | i zero or 1 time |
| i{n} | i occurs n times in sequence |
| i{n1,n2} | i occurs n1 - n2 times in sequence |
| i{n1,n2}? | non greedy match, see above example |
| i{n,} | i occures >= n times |
| [:alnum:] | Alphanumeric characters: [:alpha:] and [:digit:] |
| [:alpha:] | Alphabetic characters: [:lower:] and [:upper:] |
| [:blank:] | Blank characters: e.g. space, tab |
| [:cntrl:] | Control characters |
| [:digit:] | Digits: 0 1 2 3 4 5 6 7 8 9 |
| [:graph:] | Graphical characters: [:alnum:] and [:punct:] |
| [:lower:] | Lower-case letters in the current locale |
| [:print:] | Printable characters: [:alnum:], [:punct:] and space |
| [:punct:] | Punctuation character: ! " # $ % & ' ( ) * + , - . / : ; < = > ? @ [ \ ] ^ _ ` { | } ~ |
| [:space:] | Space characters: tab, newline, vertical tab, form feed, carriage return, space |
| [:upper:] | Upper-case letters in the current locale |
| [:xdigit:] | Hexadecimal digits: 0 1 2 3 4 5 6 7 8 9 A B C D E F a b c d e f |
subset Function
subset() function returns subsets of vectors, matrices or data frames which meet conditions.
subset(x, ...)
## Default S3 method:
subset(x, subset, ...)
## S3 method for class 'matrix'
subset(x, subset, select, drop = FALSE, ...)
## S3 method for class 'data.frame'
subset(x, subset, select, drop = FALSE, ...)
x: object to be subsetted
subset: logical expression indicating elements or rows to keep: missing values are taken as false
select: expression, indicating columns to select from a data frame
drop: passed on to [ indexing operator
...
> BOD
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
> subset(BOD,select="Time")
Time
1 1
2 2
3 3
4 4
5 5
6 7
> subset(BOD,demand<16, select="demand")
demand
1 8.3
2 10.3
5 15.6
substr Function
substr() function extract or replace substrings in a character vector.
substr(x, start, stop)
substring(text, first, last = 1000000L)
substr(x, start, stop) <- value
substring(text, first, last = 1000000L) <- value
x,text: character vector
start, first: integer, the first element to be replaced
stop, last: integer, the last element to be replaced
value: character vector, recycled if necessary
> substr("tutorial",2,3)
[1] "ut"
> x <- c("green","red","blue")
> substr(x,2,3)
[1] "re" "ed" "lu"
> substring(x,2,3)
[1] "re" "ed" "lu"
sum Function
sum() function adds up all elements of a vector.
sum(x)
x: numeric vector
> sum(1:10)
[1] 55
> sum(c(3,4,5))
[1] 12
> sum(c(2,3))
[1] 5
summary Function
summary() function is a generic function used to produce result summaries of the results of various model fitting functions. The function invokes particular methods which depend on the class of the first argument.
summary(object, ...)
## Default S3 method:
summary(object, ..., digits = max(3, getOption("digits")-3))
## S3 method for class 'data.frame'
summary(object, maxsum = 7,
digits = max(3, getOption("digits")-3), ...)
## S3 method for class 'factor'
summary(object, maxsum = 100, ...)
## S3 method for class 'matrix'
summary(object, ...)
object: R object
maxsum: interger, indicating how many levels should be shown for factors
digits: integer, used for number formatting with signif() (for summary.default) or format() (for summary.data.frame)
> x <- c("green","red","blue")
> summary(x)
Length Class Mode
3 character character
> summary(BOD)
Time demand
Min. :1.000 Min. : 8.30
1st Qu.:2.250 1st Qu.:11.62
Median :3.500 Median :15.80
Mean :3.667 Mean :14.83
3rd Qu.:4.750 3rd Qu.:18.25
Max. :7.000 Max. :19.80
svd Function
svd() function computes the singular-value decomposition of a rectangular matrix.
svd(x, nu = min(n, p), nv = min(n, p), LINPACK = FALSE)
La.svd(x, nu = min(n, p), nv = min(n, p))
x: a numeric, logical or complex matrix
nu: the number of left singular vectors to be computed. This must between 0 and n = nrow(x)
nv: the number of right singular vectors to be computed. This must be between 0 and p = ncol(x)
LINPACK: logical. Should LINPACK be used (for compatibility with R < 1.7.0)? In this case nu must be 0, nrow(x) or ncol(x)
> x <- matrix(1:16,4,4)
> x
[,1] [,2] [,3] [,4]
[1,] 1 5 9 13
[2,] 2 6 10 14
[3,] 3 7 11 15
[4,] 4 8 12 16
> svd(x)
$d
[1] 3.862266e+01 2.071323e+00 2.076990e-15 4.119458e-16
$u
[,1] [,2] [,3] [,4]
[1,] -0.4284124 -0.7186535 0.43803202 0.3288281
[2,] -0.4743725 -0.2738078 -0.82913672 -0.1119477
[3,] -0.5203326 0.1710379 0.34417739 -0.7625890
[4,] -0.5662928 0.6158835 0.04692732 0.5457086
$v
[,1] [,2] [,3] [,4]
[1,] -0.1347221 0.82574206 0.5322301 -0.1293488
[2,] -0.3407577 0.42881720 -0.6132292 0.5691660
[3,] -0.5467933 0.03189234 -0.3702319 -0.7502855
[4,] -0.7528288 -0.36503251 0.4512310 0.3104683
sweep Function
sweep() function returns an array obtained from an input array by sweeping out a summary statistic.
sweep(x, MARGIN, STATS, FUN="-", check.margin=TRUE, ...)
x: an array
MARGIN: a vector of indices giving the extent(s) of x which correspond to STATS
STATS: the summary statistic which is to be swept out
FUN: the function to be used to carry out the sweep
check.margin: logical. If TRUE (the default), warn if the length or dimensions of STATS do not match the specified dimensions of x. Set to FALSE for a small speed gain when you know that dimensions match
...
> require(stats)
> med.att <- apply(attitude, 2, median)
> med.att
rating complaints privileges learning raises critical advance
65.5 65.0 51.5 56.5 63.5 77.5 41.0
> sweep(data.matrix(attitude),2, med.att)
rating complaints privileges learning raises critical advance
[1,] -22.5 -14 -21.5 -17.5 -2.5 14.5 4
[2,] -2.5 -1 -0.5 -2.5 -0.5 -4.5 6
[3,] 5.5 5 16.5 12.5 12.5 8.5 7
[4,] -4.5 -2 -6.5 -9.5 -9.5 6.5 -6
[5,] 15.5 13 4.5 9.5 7.5 5.5 6
[6,] -22.5 -10 -2.5 -12.5 -9.5 -28.5 -7
[7,] -7.5 2 -9.5 -0.5 2.5 -9.5 -6
[8,] 5.5 10 -1.5 -1.5 6.5 -11.5 0
[9,] 6.5 17 20.5 10.5 7.5 5.5 -10
[10,] 1.5 -4 -6.5 -9.5 -1.5 2.5 0
[11,] -1.5 -12 1.5 1.5 -5.5 -10.5 -7
[12,] 1.5 -5 -4.5 -17.5 -4.5 -3.5 0
[13,] 3.5 -3 5.5 -14.5 -8.5 -14.5 -16
[14,] 2.5 18 31.5 -11.5 -4.5 -0.5 -6
[15,] 11.5 12 2.5 15.5 15.5 -0.5 5
[16,] 15.5 25 -1.5 15.5 -3.5 -23.5 -5
[17,] 8.5 20 12.5 12.5 15.5 1.5 22
[18,] -0.5 -5 13.5 18.5 -8.5 2.5 19
[19,] -0.5 5 -5.5 0.5 11.5 7.5 5
[20,] -15.5 -7 16.5 -2.5 0.5 0.5 11
[21,] -15.5 -25 -18.5 -22.5 -20.5 -13.5 -8
[22,] -1.5 -4 0.5 5.5 2.5 2.5 0
[23,] -12.5 1 0.5 -6.5 -0.5 2.5 -4
[24,] -25.5 -28 -9.5 1.5 -13.5 -20.5 8
[25,] -2.5 -11 -9.5 -8.5 2.5 -2.5 -8
[26,] 0.5 12 14.5 6.5 24.5 -1.5 31
[27,] 12.5 10 6.5 17.5 16.5 0.5 8
[28,] -17.5 -8 -7.5 -11.5 -12.5 5.5 -3
[29,] 19.5 20 19.5 14.5 13.5 -3.5 14
[30,] 16.5 17 -12.5 2.5 0.5 0.5 -2
switch Function
switch() function evaluates EXPR and accordingly chooses one of the further arguments (in ...).
switch(EXPR, ...)
EXPR: an expression evaluating to a number or a character string
...: the list of alternatives. If it is intended that EXPR has a character-string value these will be named, perhaps except for one alternative to be used as a ‘default’ value
> switch(x,6+4,mean(1:8),rnorm(4))
[1] -0.1534941 2.1080748 0.6758030 1.3047233
> switch(2,6+4,mean(1:8),rnorm(4))
[1] 4.5
> y <- switch(5,6+4,mean(1:8),rnorm(4))
> y
NULL
If the result of x is character, then the element of "..." which match the result will be executed,
if no match, return NULL.
> x <- "red"
> switch(x, red="cloth", size=5, name="table")
[1] "cloth"
> require(stats)
> centre <- function(x, type) {
+ switch(type,
+ mean = mean(x),
+ median = median(x),
+ trimmed = mean(x, trim = .1))
+ }
> x <- rcauchy(10)
> centre(x, "mean")
[1] 0.6410266
> centre(x, "median")
[1] 0.8064962
> centre(x, "trimmed")
[1] 0.8390471
Sys Function
List of Sys functions:
| Function | Description |
| Sys.chmod | Directory and file permission |
| Sys.date | Current date and time |
| Sys.getenv | Get environment Variables |
| Sys.getlocate | Query or set aspects of the locale |
| Sys.getpid | Process ID of the R session |
| Sys.glob | Wilcard expansion on file paths |
| Sys.info | Extract system and user information |
| Sys.localeconv | Details of the Numerical, Monetary Representations in the Current Locale |
| sys.on.exit | Access the function call stack |
| sys.parent | Access the function call stack |
| Sys.readlink | Read file symbolic links |
| Sys.setenv | Set or unset environment variables |
| Sys.setlocale | Query or set aspects of the locale |
| Sys.sleep | Suspend execution for a time interval |
| sys.source | Parse and evaluate expressions from a file |
| sys.status | Acess the function call stack |
| Sys.time | Current date and time |
| Sys.timezone | Time zones |
| Sys.umask | Directory and file permission |
| Sys.unsetenv | Set or unset environment variables |
| Sys.which | Finds full paths to executables |
system Function
system() function invokes the operation system command.
system(command, intern = FALSE,
ignore.stdout = FALSE, ignore.stderr = FALSE,
wait = TRUE, input = NULL, show.output.on.console = TRUE,
minimized = FALSE, invisible = TRUE)
system2(command, args = character(),
stdout = "", stderr = "", stdin = "", input = NULL,
env = character(),
wait = TRUE, minimized = FALSE, invisible = TRUE)
command: system command
intern: a logical (not NA) which indicates whether to capture the output of the command as an R character vector
ignor.stdout, ignore.stderr: whether messages written to ‘stdout’ or ‘stderr’ should be ignored
wait: whether the R interpreter should wait for the command to finish, or run it asynchronously. This will be ignored (and the interpreter will always wait) if intern = TRUE
input: if a character vector is supplied, this is copied one string per line to a temporary file, and the standard input of command is redirected to the file
show.output.on.console, minimized, invisible: arguments that are accepted on Windows but ignored on this platform, with a warning
args:arguments of the system command
stdout, stderr:where output to ‘stdout’ or ‘stderr’ should be sent. Possible values are "", to the R console (the default), NULL or FALSE (discard output), TRUE (capture the output in a character vector) or a character string naming a file
stdin:should input be diverted? "" means the default, alternatively a character string naming a file. Ignored if input is supplied
env:set environment variables
wait: a logical (not NA) indicating whether the R interpreter should wait for the command to finish, or run it asynchronously. This will be ignored (and the interpreter will always wait) if stdout = TRUE
t Function
t() function transposes a matrix or data.frame.
t(x)
x: matrix or data.frame
> BOD
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
> class(BOD)
[1] "data.frame"
> t(BOD)
[,1] [,2] [,3] [,4] [,5] [,6]
Time 1.0 2.0 3 4 5.0 7.0
demand 8.3 10.3 19 16 15.6 19.8
> x <- matrix(1:9,3,3)
> x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> class(x)
[1] "matrix"
> t(x)
[,1] [,2] [,3]
[1,] 1 2 3
[2,] 4 5 6
[3,] 7 8 9
table Function
table() function uses the cross-classifying factors to build a contingency table of the counts at each combination of factor levels.
table(..., exclude = if (useNA == "no") c(NA, NaN), useNA = c("no",
"ifany", "always"), dnn = list.names(...), deparse.level = 1)
as.table(x, ...)
is.table(x)
as.data.frame(x, row.names = NULL, ...,
responseName = "Freq", stringsAsFactors = TRUE)
...: one or more objects which can be interpreted as factors (including character strings), or a list (or data frame) whose components can be so interpreted. (For as.table and as.data.frame, arguments passed to specific methods.)
exclude: levels to remove from all factors in .... If set to NULL, it implies useNA="always"
useNA: whether to include extra NA levels in the table
dnn: the names to be given to the dimensions in the result (the dimnames names)
deparse.level: controls how the default dnn is constructed
x: an arbitrary R object, or an object inheriting from class "table" for the as.data.frame method
row.names: a character vector giving the row names for the data frame
responseName: The name to be used for the column of table entries, usually counts
stringsAsFactors: logical: should the classifying factors be returned as factors (the default) or character vectors?
> x <- matrix(1:9,3,3)
> x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> y <- as.table(x)
> y
A B C
A 1 4 7
B 2 5 8
C 3 6 9
> is.table(y)
[1] TRUE
tabulate Function
tabulate() function takes the integer-valued vector bin and counts the number of times each integer occurs in it.
tabulate(bin, nbins = max(1, bin, na.rm = TRUE))
bin: numeric vector or factor
nbins: the number of bins to be used
> tabulate(c(3,5,4))
[1] 0 0 1 1 1
> tabulate(c(3,5,4,8),nbins=4)
[1] 0 0 1 1
> tabulate(c(3,5,4,8),nbins=8)
[1] 0 0 1 1 1 0 0 1
tan Function
tan() function computes the tangent value of numeric value.
tan(x)
x: Numeric value, array or vector
> tan(pi)
[1] -1.224647e-16
> tan(pi/4)
[1] 1
> tan(0)
[1] 0
> x <- c(pi, pi/4, 0)
> tan(x)
[1] -1.224647e-16 1.000000e+00 0.000000e+00
X
(deg) |
X
(Rad) |
tangent(X) |
| 180 ̊ |
π |
0 |
| 150 ̊ |
5π/6 |
-0.57735 |
| 135 ̊ |
3π/4 |
-1 |
| 120 ̊ |
2π/3 |
-1.732051 |
| 90 ̊ |
π/2 |
Out of Range |
| 60 ̊ |
π/3 |
1.732051 |
| 45 ̊ |
π/4 |
1 |
| 30 ̊ |
π/6 |
0.57735 |
| 0 ̊ |
0 |
0 |
tanh Function
tanh() function computes the hyperbolic tangent of numberic data.
tanh(x)
x: Numeric value, array or vector.
> tanh(1)
[1] 0.7615942
> tanh(0)
[1] 0
> tanh(-2)
[1] -0.9640276
> x <- c(1,0,-2)
> tanh(x)
[1] 0.7615942 0.0000000 -0.9640276
tapply Function
tapply() applies a function to each cell of a ragged array.
tapply(X, INDEX, FUN = NULL, ..., simplify = TRUE)
•
X: vector
•
INDEX: list of one of more factors
•
FUN: the function
•
simplify: if true, return an array of scalar, other wise an array of list
...
>Orange #R built-in dataset, Growth of Orange Trees
Tree age circumference
1 1 118 30
2 1 484 58
3 1 664 87
4 1 1004 115
5 1 1231 120
6 1 1372 142
7 1 1582 145
8 2 118 33
9 2 484 69
10 2 664 111
11 2 1004 156
12 2 1231 172
13 2 1372 203
14 2 1582 203
15 3 118 30
16 3 484 51
17 3 664 75
18 3 1004 108
19 3 1231 115
20 3 1372 139
21 3 1582 140
22 4 118 32
23 4 484 62
24 4 664 112
25 4 1004 167
26 4 1231 179
27 4 1372 209
28 4 1582 214
29 5 118 30
30 5 484 49
31 5 664 81
32 5 1004 125
33 5 1231 142
34 5 1372 174
35 5 1582 177
Calculate the mean circumference of different Tree groups:
> tapply(Orange$circumference,Orange$Tree,mean)
3 1 5 2 4
94.00000 99.57143 111.14286 135.28571 139.28571
Return a list:
> tapply(Orange$circumference,Orange$Tree,mean,simplify=FALSE)
$`3`
[1] 94
$`1`
[1] 99.57143
$`5`
[1] 111.1429
$`2`
[1] 135.2857
$`4`
[1] 139.2857
tempfile Function
tempfile() function returns a vector of character strings which can be used as names for temporary files.
tempfile(pattern = "file", tmpdir = tempdir(), fileext = "")
tempdir()
pattern: non-empty character vector giving the initial part of the name
tmpdir: non-empty character vector giving the directory name
fileext: non-empty character vector giving the file extension
> tempdir()
[1] "C:\\Users\\...\\AppData\\Local\\Temp\\Rtmpspq3L1"
> tempfile("tp")
[1] "C:\\Users\\...\\AppData\\Local\\Temp\\Rtmpspq3L1\\tp63ec15e91ffc"
> tempfile("tp",fileext=".csv")
[1] "C:\\Users\\...\\Temp\\Rtmpspq3L1\\tp63ec522f50cd.csv"
> tempfile("tp",fileext=c(".csv",".txt"))
[1] "C:\\Users\\...\\Temp\\Rtmpspq3L1\\tp63ec2bf25694.csv"
[2] "C:\\Users\\...\\Temp\\Rtmpspq3L1\\tp63ec2b4729a8.txt"
textConnection Function
textConnection() function inputs and outputs text connections.
textConnection(object, open = "r", local = FALSE,
encoding = c("", "bytes", "UTF-8"))
textConnectionValue(con)
object: character. A description of the connection. For an input this is an R character vector object, and for an output connection the name for the R character vector to receive the output, or NULL (for none)
open: character. Either "r" (or equivalently "") for an input connection or "w" or "a" for an output connection
local: logical. Used only for output connections. If TRUE, output is assigned to a variable in the calling environment. Otherwise the global environment is used
encoding: character. Used only for input connections. How marked strings in object should be handled: converted to the current locale, used byte-by-byte or translated to UTF-8
con: an output text connection
> letters
[1] "a" "b" "c" "d" "e" "f" "g" "h" "i" "j" "k" "l" "m" "n"
"o" "p" "q" "r" "s"
[20] "t" "u" "v" "w" "x" "y" "z"
> x <- textConnection(letters)
> readLines(x, 2)
[1] "a" "b"
> close(x)
tolower Function
tolower() function converter string to its lower case.
tolower(x)
x: character vector
> tolower("EndMemo R Tutorial")
[1] "endmemo r tutorial"
> x <- c("Green", "Red", "Black")
> tolower(x)
[1] "green" "red" "black"
toString Function
toString() function produces a single character string describing an R object.
toString(x, ...)
toString(x, width = NULL, ...)
x: R object
width: Suggestion for the maximum field width. Values of NULL or 0 indicate no maximum. The minimum value accepted is 6 and smaller values are taken as 6
...
> x <- c("Green", "Red", "Black")
> toString(x)
[1] "Green, Red, Black"
> toString(x,width=5)
[1] "Gr...."
> toString(x,width=12)
[1] "Green, R...."
toupper Function
toupper() function converts a string to its upper case.
toupper(x)
x: character vector
> x <- c("Green", "Red", "Black")
> toupper(x)
[1] "GREEN" "RED" "BLACK"
trace Function
trace() function allows user to insert debugging code at chosen
places in any function.
trace(what, tracer, exit, at, print, signature,
where = topenv(parent.frame()), edit = FALSE)
untrace(what, signature = NULL, where = topenv(parent.frame()))
tracingState(on = NULL)
.doTrace(expr, msg)
what: the name (quoted or not) of a function to be traced or untraced. For untrace or for trace with more than one argument, more than one name can be given in the quoted form, and the same action will be applied to each one
tracer: either a function or an unevaluated expression. The function will be called or the expression will be evaluated either at the beginning of the call, or before those steps in the call specified by the argument at
exit: either a function or an unevaluated expression. The function will be called or the expression will be evaluated on exiting the function
at: optional numeric vector or list. If supplied, tracer will be called just before the corresponding step in the body of the function
print: if TRUE (as per default), a descriptive line is printed before any trace expression is evaluated
signature: if this argument is supplied, it should be a signature for a method for function what. In this case, the method, and not the function itself, is traced
edit: gor complicated tracing, such as tracing within a loop inside the function, you will need to insert the desired calls by editing the body of the function. If so, supply the edit argument either as TRUE, or as the name of the editor you want to use. Then trace() will call edit and use the version of the function after you edit it
where: where to look for the function to be traced; by default, the top-level environment of the call to trace
on: logical; a call to the support function tracingState returns TRUE if tracing is globally turned on, FALSE otherwise. An argument of one or the other of those values sets the state. If the tracing state is FALSE, none of the trace actions will actually occur (used, for example, by debugging functions to shut off tracing during debugging)
expr,msg: arguments to the support function .doTrace, calls to which are inserted into the modified function or method: expr is the tracing action (such as a call to browser(), and msg is a string identifying the place where the trace action occurs
> trace(print)
> for(i in 1:5) print(i)
trace: print(i)
[1] 1
trace: print(i)
[1] 2
trace: print(i)
[1] 3
trace: print(i)
[1] 4
trace: print(i)
[1] 5
> untrace(print)
> for(i in 1:5) print(i)
[1] 1
[1] 2
[1] 3
[1] 4
[1] 5
transform Function
transform() function converts its first arguments to a data frame if possible.
transform('_data',...)
_data: R object to be transformed
...
> BOD
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
> transform(BOD, demand = -demand)
Time demand
1 1 -8.3
2 2 -10.3
3 3 -19.0
4 4 -16.0
5 5 -15.6
6 7 -19.8
try Function
try() function is a wrapper to run an expression that might fail and allow the user's code to handle error-recovery.
try(expr, silent=FALSE)
tryCatch(expr, error=function(e) e)
expr: R expression
silent: logical: should the report of error messages be suppressed?
error: error handling function
> x <- 3
> try(x > 5)
[1] FALSE
> tryCatch(x>5,error=print("error"))
[1] "error"
[1] FALSE
t test
t.test(...) function returns a t test result of two group data sets. It's expression is:
t.test(x, y = NULL,
alternative = c("two.sided", "less", "greater"),
mu = 0, paired = FALSE, var.equal = FALSE,
conf.level = 0.95, ...)
x,y:Numeric vectors
alternative:Alternativ Hypothesis
mu:True value of the mean
paired:Paired t-test or not
...
Suppose we have two dataset, let's do a t test
>x <- c(1.2,3.4,1.3,-2.1,5.6,2.3,3.2,2.4,2.1,1.8,1.7,2.2)
>y <- c(2.4,5.7,2.0,-3,13,5,6.2,4.8,4.2,3.5,3.7,5.2)
>ret <- t.test(x,y)
>ret
Welch Two Sample t-test
data: x and y
t = -1.9667, df = 15.943, p-value = 0.06688
alternative hypothesis: true difference in means is not equal to 0
95 percent confidence interval:
-4.7799367 0.1799367
sample estimates:
mean of x mean of y
2.091667 4.391667
type Function
type() function determines the type or storage mode of an R object.
typeof(x)
x: R object
> x <- 3
> typeof(x)
[1] "double"
> x <- c(3,4,5)
> typeof(x)
[1] "double"
> BOD
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
> typeof(BOD)
[1] "list"
unique Function
unique() function removes duplicated elements/rows from a vector, data frame or array.
unique(x, incomparables = FALSE, ...)
unique(x, incomparables = FALSE, fromLast = FALSE, ...)
unique(x, incomparables = FALSE, MARGIN = 1,
fromLast = FALSE, ...)
unique(x, incomparables = FALSE, MARGIN = 1,
fromLast = FALSE, ...)
x: vector, data frame, array or NULL
incomparables: a vector of values that cannot be compared. FALSE is a special value, meaning that all values can be compared, and may be the only value accepted for methods other than the default. It will be coerced internally to the same type as x
fromLast: logical indicating if duplication should be considered from the last, i.e., the last (or rightmost) of identical elements will be kept. This only matters for names or dimnames
...: arguments for particular methods
MARGIN: the array margin to be held fixed: a single integer
> x <- c(2:8,4:10)
> x
[1] 2 3 4 5 6 7 8 4 5 6 7 8 9 10
> unique(x)
[1] 2 3 4 5 6 7 8 9 10
unlink Function
unlink() function deletes a file or directory.
unlink(x, recursive = FALSE)
x: file or directory
recursive: whether directories be deleted recursively
unlist Function
unlist(x) function simplifies a list to produce a vector which contains all the atomic components which occur in x.
unlist(x, recursive = TRUE, use.names = TRUE)
x: list or vector
recursive: logical, should unlisting be applied to list components of x
use.names: logical, should names be preserved
> BOD
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
> is.list(BOD)
[1] TRUE
> unlist(BOD)
Time1 Time2 Time3 Time4 Time5 Time6 demand1
1.0 2.0 3.0 4.0 5.0 7.0 8.3
demand2 demand3 demand4 demand5 demand6
10.3 19.0 16.0 15.6 19.8
> unlist(BOD,use.names=FALSE)
[1] 1.0 2.0 3.0 4.0 5.0 7.0 8.3 10.3 19.0 16.0 15.6 19.8
unname Function
unname() function removes the names or dimnames attribute of an R object.
unname(obj, force=FALSE)
obj: an R object
force: logical; if true, the dimnames (names and row names) are removed even from data.frames
> BOD
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
> unname(BOD)
NA NA
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
Vector Data Type
R vector data type is similar to array of other programming languages. It's consisted of an ordered number of elements.
The elements can be numeric (integer, double), logical, character, complex, or raw.
Vector assignment:
>v <- c(2,3,5.5,7.1,2.1,3)
>v
[1] 2.0 3.0 5.5 7.1 2.1 3.0
Other vector assignment syntax:
>assign("v",c(2,3,5.5,7.1,2.1,3))
>c(2,3,5.5,7.1,2.1,3) -> v
The 1st element of R vector is indexed as 1, not 0 as some other programming languages.
Access the 3rd elements of vector v:
>v[3]
[1] 2
R can operate vector like a single element. e.g.
>1/v
[1] 0.5000000 0.3333333 0.1818182 0.1408451 0.4761905 0.3333333
>2+v
[1] 4.0 5.0 7.5 9.1 4.1 5.0
>v2 <- v + 1/v + 5
>v2
[1] 7.500000 8.333333 10.681818 12.240845 7.576190 8.333333
Judge a data structure is vector or not:
>is.vector(v)
[1] TRUE
>is.vector(3,mode="any")
[1] TRUE
>is.vector(3,mode="list")
[1] FALSE
Under default mode "any", logical, number, character are treated as vectors with length 1. It will retrun FALSE only if the object
being judged has name attribute. Under mode "numeric", is.vector will return true for vectors of types integer or double.
and mode "integer" can only be true for vectors of type integer.
Other methods for generating regular vectors:
>v <- 1:10
>v
[1] 1 2 3 4 5 6 7 8 9 10
>v <- rep(2,10)
>v
[1] 2 2 2 2 2 2 2 2 2 2
>v <- seq(1,5,by=0.5)
>v
[1] 1.0 1.5 2.0 2.5 3.0 3.5 4.0 4.5 5.0
>v <- seq(length=10,from=1,by=0.5)
>v
[1] 1.0 1.5 2.0 2.5 3.0 3.5 4.0 4.5 5.0 5.5
version Function
version() function provides detailed information about current R used.
R.Version()
R.version
R.version.string
version
getRversion()
> version
_
platform x86_64-w64-mingw32
arch x86_64
os mingw32
system x86_64, mingw32
status
major 3
minor 0.1
year 2013
month 05
day 16
svn rev 62743
language R
version.string R version 3.0.1 (2013-05-16)
nickname Good Sport
> getRversion()
[1] ‘3.0.1’
> R.Version()
$platform
[1] "x86_64-w64-mingw32"
$arch
[1] "x86_64"
$os
[1] "mingw32"
$system
[1] "x86_64, mingw32"
$status
[1] ""
$major
[1] "3"
$minor
[1] "0.1"
$year
[1] "2013"
$month
[1] "05"
$day
[1] "16"
$`svn rev`
[1] "62743"
$language
[1] "R"
$version.string
[1] "R version 3.0.1 (2013-05-16)"
$nickname
[1] "Good Sport"
warning Function
warning() function generates a warning message that corresponds to its argument(s) and (optionally) the expression or function from which it was called.
warning(..., call. = TRUE, immediate. = FALSE, domain = NULL)
suppressWarnings(expr)
warnings(...)
...: zero or more objects which can be coerced to character (and which are pasted together with no separator) or a single condition object
call.: logical, indicating if the call should become part of the warning message
immediate.: logical, indicating if the call should be output immediately, even if getOption("warn") <= 0
expr: expression to evaluate
domain: If NA, messages will not be translated
which Function
which() function gives the TRUE indices of a logical object, allowing for array indices.
which(x, arr.ind = FALSE, useNames = TRUE)
arrayInd(ind, .dim, .dimnames = NULL, useNames = FALSE)
x: logical vector or array. NAs are allowed and omitted (treated as if FALSE)
arr.ind: logical; should array indices be returned when x is an array?
ind: integer-valued index vector, as resulting from which(x)
.dim: integer vector
.dimnames: optional list of character dimnames(.), of which only .dimnames[[1]] is used
useNames: logical indicating if the value of arrayInd() should have (non-null) dimnames at all
> which(letters=="h")
[1] 8
> BOD
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
> which(BOD$demand == 16)
[1] 4
> x <- matrix(1:9,3,3)
> x
[,1] [,2] [,3]
[1,] 1 4 7
[2,] 2 5 8
[3,] 3 6 9
> which(x %% 3 == 0, arr.ind=TRUE)
row col
[1,] 3 1
[2,] 3 2
[3,] 3 3
> which(x %% 3 == 0, arr.ind=FALSE)
[1] 3 6 9
while Loop
while() loop will execute a block of commands until the condition is no longer satisfied.
while(cond) expr
cond: condition
expr: expression
> x <- 1
> while(x < 5) {x <- x+1; print(x);}
[1] 2
[1] 3
[1] 4
next can skip one step of the loop.
break will end the loop abruptly.
Let's break the loop when x=3:
> x <- 1
> while(x < 5) {x <- x+1; if (x == 3) break; print(x); }
[1] 2
Let's skip one step when x=3:
> x <- 1
> while(x < 5) {x <- x+1; if (x == 3) next; print(x);}
[1] 2
[1] 4
[1] 5
wilcoxon rank test
wilcox.test() function performs wilcoxon rank test, which assumes that the means of two unnormally distributed datasets are equal.
wilcox.test(x, ...)
wilcox.test(x, y, alternative = c("two.sided", "less", "greater"),
mu = 0, paired = FALSE, exact = NULL, correct = TRUE,
conf.int=FALSE, conf.level = 0.95, ...)
x,y: Unnormally distributed data sets
ratio: Hypothesized ratio of x/y, default is 1
alternative: alternative hypothesis, including "two.sided","greater","less"
conf.level: confidence level
...
- c(1,5,9,24,56,21,3,7,21,4)
> y <- c(12,15,5,9,9,14,56,22,3,7,32,5)
> wilcox.test(x,y)
Wilcoxon rank sum test with continuity correction
data: x and y
W = 51.5, p-value = 0.5966
alternative hypothesis: true location shift is not equal to 0
Warning message:
In wilcox.test.default(x, y) : cannot compute exact p-value with ties
Since the p-value = 0.5966 is much higher than 0.05, the hypothesis that the two means are equal is accepted.
> y <- c(1233,4356,987,39999,1111,200000)
> wilcox.test(x,y)
Wilcoxon rank sum test with continuity correction
data: x and y
W = 0, p-value = 0.001364
alternative hypothesis: true location shift is not equal to 0
Warning message:
In wilcox.test.default(x, y) : cannot compute exact p-value with ties
p-value = 0.001363 which is much lower than 0.05, rejects the hypothesis.
with Function
with() function evaluates an R expression in an environment constructed from data, possibly modifying the original data.
with(data, expr, ...)
within(data, expr, ...)
data: data to use for constructing an environment. For the default with method this may be an environment, a list, a data frame, or an integer as in sys.call. For within, it can be a list or a data frame
expr: expression
...
> BOD
Time demand
1 1 8.3
2 2 10.3
3 3 19.0
4 4 16.0
5 5 15.6
6 7 19.8
> with(BOD,{BOD$demand <- BOD$demand + 1; print(BOD$demand)})
[1] 9.3 11.3 20.0 17.0 16.6 20.8
> within(BOD,{BOD$demand <- BOD$demand + 1; print(BOD$demand)})
[1] 9.3 11.3 20.0 17.0 16.6 20.8
Time demand BOD.Time BOD.demand
1 1 8.3 1 9.3
2 2 10.3 2 11.3
3 3 19.0 3 20.0
4 4 16.0 4 17.0
5 5 15.6 5 16.6
6 7 19.8 7 20.8
withVisible Function
withVisible() function evaluates an expression, returning it in a two element list containing its value and a flag showing whether it would automatically print.
withVisible(x)
x: expression to be evalutated
> x <- 3
> withVisible(x <- 1)
$value
[1] 1
$visible
[1] FALSE
> x <- c(3,4,2,1,7,56)
> withVisible(x <- 10)
$value
[1] 10
$visible
[1] FALSE
> x <- c(3,4,2,1,7,56)
> withVisible(x < 10)
$value
[1] TRUE TRUE TRUE TRUE TRUE FALSE
$visible
[1] TRUE
Write Data to File
write(...) function writes data to a file. It's usage is:
write(x, file = "data",
ncolumns = if(is.character(x)) 1 else 5,
append = FALSE, sep = " ")
x: The data to be written, including vector, matrix, scalar, but not data frame
file: Out file name, if empty, then print to the screen
ncolumns: Number of columns in the file
append: Append model, otherwise new file will be created
sep: Seperator, default is " "
Let's see a matrix example (we will print to the screen, you may give a file name for writing to the file):
>x <- matrix(c(3,5,7,1,9,4),nrow=3,ncol=2,byrow=TRUE)
>write(x,"")
3 7 9 5 1
4
>write(x,"",ncolumns=2,sep=",")
3,7
9,5
1,4
write.table(...) write data frame to a file.
write.table(x, file = "", append = FALSE, quote = TRUE, sep = " ",
eol = "\n", na = "NA", dec = ".", row.names = TRUE,
col.names = TRUE, qmethod = c("escape", "double"),
fileEncoding = "")
Let's write the data set BOD to the screen:
>write.table(BOD,"",sep=",")
"Time","demand"
"1",1,8.3
"2",2,10.3
"3",3,19
"4",4,16
"5",5,15.6
"6",7,19.8

write.csv has similar function with
write.table.
write.table function
write.table(...) function writes a matrix or data frame into a file. It's usage is:
write.table(x, file = "", append = FALSE, quote = TRUE, sep = " ",
eol = "\n", na = "NA", dec = ".", row.names = TRUE,
col.names = TRUE, qmethod = c("escape", "double"),
fileEncoding = "")
write.csv(x, file = "", append = FALSE, quote = TRUE, sep = " ",
eol = "\n", na = "NA", dec = ".", row.names = TRUE,
col.names = TRUE, qmethod = c("escape", "double"),
fileEncoding = "")
write.csv2(...)
x: The matrix or data frame to be written
file: Out file name, if empty, then print to the screen
append: Append model, otherwise new file will be created
sep: Seperator, default is " "
...
Let's see a matrix example (we will print to the screen, you may give a file name for writing to the file):
> x <- matrix(c(3,5,7,1,9,4),nrow=3,ncol=2,byrow=TRUE)
> write.table(x,"")
"V1" "V2"
"1" 3 5
"2" 7 1
"3" 9 4
Let's write into a file and use "," as field separator:
> write.table(x,"test.csv",sep=",")
The content of "test.csv" is:
"V1","V2"
"1",3,5
"2",7,1
"3",9,4
Z-test
Formula for Z Score:
z = √n(x - x0)/σ
Where:
n: Sample number
x: Population mean
x0: Hyposized population mean
σ: Standard Deviation
We hypothesize water volume will not change under X rays. So we checked 100 bottles of drinking water with 300 ml volume,
and recorded the volume difference from 300 ml.
We will test the Hypothesis H0: σ = 0 against σ ≠ 0.
Data in "tp.txt":
Calculate the Z Score:
> x <- read.csv("tp.txt",header=F)
> x <- x[1:100,]
> z <- sqrt(100) * (mean(x) - 0)/sd(x)
> z
[1] -0.2334861
Calculate P value:
> p <- 2 * pnorm(-abs(z),0,1)
> p
[1] 0.8153839
Since p>0.05, we accept the hypothesis.
We then hypothesize water volume will not change under higher temperature at 80 degrees. So we checked 100 bottles of drinking water with 300 ml volume,
and recorded the volume difference from 300 ml.
We will test the Hypothesis H
0: σ = 0 against σ ≠ 0.
Data in "tp.txt":
Calculate the Z Score:
> x <- read.csv("tp.txt",header=F)
> x <- x[1:100,]
> z <- sqrt(100) * (mean(x) - 0)/sd(x)
> z
[1] 18.28636
Calculate P value:
> p <- 2 * pnorm(-abs(z),0,1)
> p
[1] 1.06279e-74
Since p < 0.05, the hypothesis is rejected.












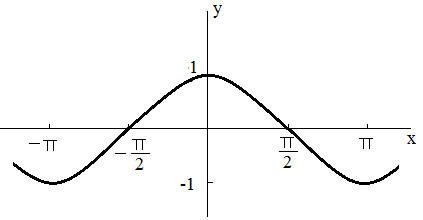










 See
See 























